Introduction
Liver cancer (LC) is a critical cause of mortality induced by cancer in humans, and in this category, hepatocellular carcinoma (HCC) is the most common subtype, accounting for about 90% of primary LCs [1]. This type of carcinoma is estimated to cause more than 800 million deaths per year [2], being the third principal reason for cancer-related death globally, and mortality levels remain high. Over the years, several treatment approaches have been tried and performed to improve therapeutic efficacy, including tumor resection, microwave ablation, and chemotherapy [3]. Risk factors for this cancer include hepatitis virus type C, type B, and cirrhosis [1, 4]. The level of hepatic damage and the cancer stage decide the prognosis of LC patients, which is not frequently at an acceptable level [5, 6]. In the first stage of HCC, surgery is practical and valuable for curative therapy [7, 8]; however, the 5-year survival rate for LC patients is unsatisfactory. Therefore, novel treatment strategies are required to reverse this unacceptable condition of disease rates in terms of high frequency, recurrence, and metastasis [9, 10].
MicroRNAs (miRs) are small RNA oligonucleotides with a length of 24-28 nucleotides. Many studies emphasize how the miR-mediated silencing complex controls gene expression [11, 12]. Briefly, miRs are transcribed by RNA polymerase II enzyme to primarymiR that can be cleaved by the Drosha enzyme in the nucleus [13]. These miRs are then processed into precursor miR. Then, by Exportin 5, the precursor miR is exported to the cytoplasm and processed by the Dicer enzyme to produce a duplex miR [14]. The mature miR is formed and binds to an Ago protein and is incorporated into the RNA-induced silencing (RISC) complex. miRs act according to the base pair relationship with their complementary sequences within the mRNA molecules. So, precursor miR has two components, miR-5p and miR-3p, and depending on the organ, both can become functional [15]. They regulate the expression of downstream targets by binding with the 3’-untranslated region (3’-UTR) of the target mRNA. Finally, mature miR causes translational repression and degradation of target mRNA [16]. Cancer treatment via inducing degradation of a specific oncogene mRNA is adversely associated with HCC initiation and development [17]. Single miRs often target multiple miRs and a single transcript may be targeted by several miRs due to their sequences. mRNAs that act as tumor suppressors (TS) are genes that play a role in DNA repair, apoptosis, and detoxification. Therapeutic miRs can function via positively regulating TS gene expression or reducing oncogene expression [18]. Another category is unregulated miRs, which may impede transmission in cell cycle irregularities and reduce miRs in HCC [1, 19]. miRs can act as TS and oncogenes. Suppressors of the oncogenic targets usually are regulated in cancer tissues. Through the induction of different pathways, biological processes play a pivotal role in cancer-related processes.
The dual role of TS and oncogenes has been reported in many studies and recognized as the target genes of miR. The difficulty of miR prediction and their biological approval is an obstacle to their therapeutic application. Because of low expression and sustainability, there is slow progress in identifying miR targets. With artificial intelligence and the advancement of biological science, computational algorithms have been created to identify miR targets [20]. We could develop such algorithms with the principles of miR-target-based diagnosis. These algorithms are used to predict many of the features of mammalian use [21]. People with alcohol or fatty liver are more at risk of HCC, which is accompanied by inflammation, necrosis, fibrosis, and continuous liver cirrhosis [22]. In addition to these risk factors, other risk factors are aflatoxin, aging, and obesity. Therefore, it is crucial to study and identify diagnostic and therapeutic targets for HCC [23].
Function of tumor suppressor miRs in hepatocellular carcinoma
There is mounting evidence that miRs might function as oncogenes or TS by directly or indirectly controlling the outflow of essential proteins involved in disease-related pathways [24, 25]. miRs affect all physiological processes, which can lead to liver diseases [26]. HCC progression has occurred as a result of impaired miRs as fundamental controllers of liver functions, for example, in conditions such as viral hepatitis, fibrosis, and HCC [27]. A reduction in the expression level of miRs in HCC may indicate that some of them may act as putative TS genes [28, 29]. TS miRs cause cell cycle arrest, apoptosis enhancement, and tumor progression reduction. miRs may function by direct or indirect control of the expression of binding proteins found in HCC-related cascades [24, 30]. Up-regulation of oncogene miRs and down-regulation of TS miRs stimulate HCC carcinogenesis through different mechanisms discussed in detail in the following subsections (Fig. 1, Table 1).
Fig. 1
The tumor suppressor (TS) genes and oncogenes have critical roles in hepatocellular carcinoma (HCC) carcinogenesis. Dysregulation of both oncogene and TS microRNAs (miRs) is involved in the pathogenesis of HCC. A number of miRs target TS and oncogenes and have shown potential for HCC diagnosis and therapy
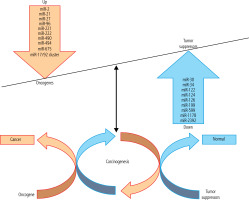
Table 1
Tumor suppressor and oncogene miRNAs and their targets and expression status in hepatocellular carcinoma
Function of miR-124
Mounting evidence has shown that the expression of miR-124 decreases in HCC [31, 32]. The exact role of miR-124 in HCC remains to be understood. To date, various target genes for the activity of this miR have been identified. Among them, the alpha catalytic subunit of phosphoinositide 3-kinase (PIK3CA) is a new target of miR-124 in the HepG2 cell line. In this regard, upregulation of miR-124 causes inhibition of PIK3CA expression, and overexpression of miR-124 significantly causes cell cycle arrest at the G1 stage. Similarly, inhibition of PIK3CA suppresses cell proliferation, while upregulation of this kinase reverses the inhibitory function of miR-124. Mechanical studies have shown that miR-124-mediated reduction of PIK3CA subsequently suppresses the PI3K/protein kinase B (AKT) pathway. In addition, miR-124 overexpression suppresses tumor growth in xenograft transplant animal models. Lang et al. found that miR-124 acts as a growth suppressor miR and plays a pivotal role in inhibiting HCC by targeting PIK3CA [33]. The miR-124 level decreased in HCC tissues, and extrauterine expression of miR-124 prevented the proliferation and migration of HCC cells. Study of aquaporin 3 (AQP3) in HCC indicated that this protein is the direct target of miR-124. AQP3 was overexpressed in HCC tissues and was inversely associated with miR-124 levels. Upregulation of miR-124 decreased AQP3 protein level, and overexpression of AQP3 induced cell proliferation and migratory ability, while knockdown of miR-124 suppressed cell proliferation and metastasis. In this study, HIPK3 circular RNA (circHIPK3) acts as a miR-124 sponge and regulates the expression of AQP3 [34]. This RNA was overexpressed in HCC tissues and was positively associated with AQP3 expression. Thus, circHIPK3 suppression inhibited cell proliferation and invasion by reducing AQP3 expression. In addition, suppression of miR-124 decreased the effect of knockHIPK3 on cell invasion and inhibited AQP3 expression [34].
Function of miR-126
Research has shown that miR-126 is deregulated and has a critical function in different cancers such as HCC, and detailed mechanisms remain to be elucidated [35]. In HCC cells and tissues, the miR-126 expression level is reduced. Low expression levels of miR-126 were correlated with poor prognosis and recurrence. Overexpression of miR-126 significantly reduced cell proliferation and migratory ability and promoted apoptosis of HCC cells [36]. Functional studies have shown that the ectopic expression of miR-126 substantially inhibits HCC-associated properties in both in vitro and animal models. Down-regulation of miR-126 has an essential role in HCC and suggests the application of miR-126-based approaches in the prognosis and treatment of HCC [37, 38].
Function of miR-122
miR-122 is a highly expressed miR in the hepatic tissue [39, 40]. It mainly regulates the metabolism of fatty acids and the regulation of liver growth, differentiation, and polyploidy of hepatocytes in several biological processes, such as homeostasis and metabolisms [35, 41, 42]. Among these miRs, miR-122 is critically essential in HCC [43, 44]. The liver-specific miR-122 is the most expressed miR in the liver and modulates hepatocyte functions [45]. The HCC cells expressed miR-122, and exaggerated expression of this miR induced apoptosis and suppressed the proliferation of HCC cells [46]. The function of miR-122 in LC was investigated by transfection of miR-122 mimic in mice. It identified that this miR has TS function, and previous studies highlighted its pivotal role in hepatic metabolism and hepatocyte proliferation [47, 48]. Cyclin G1 is a direct target of miR-122, and the miR-122/cyclin G1 interplay mediates p53 function and affects chemosensitivity in HCC cells. In an animal model of HCC, the lack of cyclin G1 is correlated with less tumor development [49, 50]. These results show that reducing the expression of this miR, as one of the most important TS, plays a pivotal role in the occurrence of LC. miR-122 reduces the migratory ability of the cells by suppressing the synthesis of tumor necrosis factor-converting enzyme. Thus, decreased miR-122 expression is linked to the presence of metastases and a faster time to recurrence in individuals with LC [51].
Function of miR-199 family
miR-199 has a crucial function in the progression of chronic liver damage to fibrosis and cirrhosis [52]. This miR is down-regulated in HCC compared to non-tumor samples and connected to the regulation of mTOR, cMet, HIF-1, and CD44. miR-199a/b-5p acts as an HCC-specific TS, which inhibits ROCK1 and regulates ROCK1/MLC and PI3K/AKT cascades, which are crucial for HCC development. A potential function of miR-199 as an HCC biomarker has been suggested [43, 53, 54]. The members of the miR-199 family are frequently down-regulated in HCC. miR-199a is a highly conserved miR that is not continuously regulated in many human cancers [55]. Microarray analysis showed that miR-199a is not primarily regulated in HCC. miR-199a expression levels in HCC tissues and adjacent tissues, HCC cell lines, and L02 normal liver cell lines were analyzed by real-time quantitative PCR techniques. miR-199a showed a significant decrease in HCC. miR199a expression levels were also significantly reduced in several human cancers when compared with adjacent tissues [56]. Li et al. assessed the miR-199b and JAG1 expression levels in 45 human tissues and HCC paracancerous tissue samples using a laboratory study at the protein level and the Transwell method to monitor the migratory ability and cell invasion [57]. The miR-199b showed a reduction in paired tumor tissues compared to the corresponding para-carcinoma tissues. miR-199b expression was closely correlated with HCC stage and size, and 5-year overall survival. The findings obtained by the Transwell assay showed that miR-199b inhibited HCC migration and cell invasion. The results indicated that miR-199b is directly associated with the 3’-UTRs of JAG1 expression level and negatively regulates JAG1 expression in HCC. These results show that miR-199b has a suppressive function and can be used as a new therapeutic marker of HCC. Also, miR-199b has TS functions in HCC by targeting JAG1 and may be a potential target treatment for LC [58].
Function of miR-30
miR-30a-3p expression level decreased in HCC cell lines compared to the LO2 normal liver cell line [59]. Elevated miR-30a-3p levels significantly suppressed MHCC-97H proliferation and migration. miR-30a-3p also inhibited tumor extension in vivo using HCC animal models. With some mechanistic experiments, COX-2 was discovered as a target of miR-30a-3p in HCC cells. Increased miR-30a-3p expression decreased COX-2 transcription levels in HCC cells, while COX-2 genetic expression could reverse the growth suppression effects of HCC cells by miR-30a-3p. In addition, using a COX-2 inhibitor, celecoxib could facilitate the anti-metastatic effects of miR-30a-3p in HCC cells [59]. Finally, a decrease in COX-2 protein levels affects the production of PGE2, which leads to a reduction in the expression of Bcl-2, caspase-3, MMP-2, and MMP-9 but increases the expression of Bax and E-cadherin, which in turn leads to higher rates of apoptosis and increases the migratory ability of the cells. Overall, miR-30a-3p can be a target for treating several cancer-associated properties, including hepatic carcinoma cell progression [59–61].
Function of miR-2392
Sun et al. assessed RNA sequencing of HCC samples and evaluated them in comparison with paired non-tumor tissues to identify the effect of miR-2392 on LC [62]. Also, the assessment of serum and tissue samples of HCC patients revealed that the expression of miR-2392 was significantly reduced compared to adjuvant tissues, which was more compatible with RNA sequencing findings. miR-2392 could dramatically inhibit the growth and migratory ability of HCC cells in vitro. Also, miR-2392 might have a TS function in this cancer. Circulating tumor cells (CTCs) disseminated in the blood can suppress shear stress-induced apoptosis in blood. Therefore, the surviving CTCs may be critical factors in migration and metastasis. Probably, the tumor microenvironment could provide indications for describing the survival mechanism of CTCs. The amplification and culture of CTCs in vitro are necessary due to the low number of CTCs. A recent study using a three-dimensional cell culture system identified a correlation between miR-2392 and induced cell spheroid formation ability [62]. Also, there was a negative correlation between the level of miR-2392 in serum and the number of CTC spheroids. These results suggest that miR-2392 might be one of the essential elements affecting the survival of CTCs [62].
Function of miR-1178
Pan et al. assessed the function of miR-1178-3p in HCC metastasis and proposed possible mechanisms. They found that miR-1178-3p was down-regulated, while TBL1XR1 was up-regulated in HCC tumors by bioinformatic study and RT-PCR experiments [63]. Also, they established the association of miR-1178-3p and TBL1XR1 utilizing a dual-luciferase reporter (DLR) assay. Moreover, gain- and loss-of-function tests revealed that overexpression of miR-1178-3p blocked the growth and migratory ability of HCC cells and decreased the xenograft tumor development and angiogenesis by modulating the decreased the xenograft tumor development and angiogenesis by modulating the TBL1XR1/PI3K/AKT pathway. They suggested that miR-1178-3p acts as a TS in HCC by targeting this axis. These discoveries could provide a therapeutic target for LC treatment [63].
Function of miR-34
Genetic factors and epigenetic mechanisms regulate the stemness of cancer stem cells (CSCs) [64, 65]. miR-34a, a TS miR and a member of the miR-34 family, was recognized as an essential regulator of p53 activity [66]. Its expression is dysregulated and down-regulated in diverse malignancies. By interfering with stemness elements, miR-34a epigenetically and negatively controls the underlying effects of CSCs [67]. Hong et al. investigated the mechanism of action of MRX34, a liposomal miR-34a mimic [68]. It has been identified that in different clinical environments, miR-34a can affect the features of CSCs and p53-mutant tumors to combat chemoresistance. Also, miR-34a has been involved in determining the characteristics of CSCs in different malignancies [67]. According to Sun et al., HCC correlates with inflammatory states of the liver, and autophagy has been shown to prevent cancer in the advanced stage by suppressing inflammation. In addition, analysis of molecular mechanisms of autophagy and inflammation can help design new therapy options. miR-34a has autophagy induction and inflammation-suppressing activity in HCC [69]. Epigenetic regulation is altered and disrupted in cancer condition as characterized by global DNA hypomethylation, leading to genomic instability and silencing specific TS genes and miRs. Sun et al. screened TET1 as a TS in HCC by bioinformatics study. The following miR microarray study demonstrated that miR-34a is a downstream miR of TET1, and its down-regulation in HCC is attributed to DNA hypermethylation [69].
Function of miR-599
Earlier miR microarray evidence revealed that hsa-miR-599 has a reduced expression level in HCC [70]. However, the process and subsequent functions of hsamiR-599 on HCC are not well understood. Tian et al. investigated the expression level of hsa-miR-599 in HCC by qRT-PCR [70]. Strangely, hsa-miR-599 had reduced expression in the investigated HCC samples.The MTT assay, wound healing, and transwell examination considered cell growth, movement, and metastasis, and the outcomes showed that over-expression of hsa-miR-599 restrained HCC cell growth and migration in vitro. Likewise, in a double luciferase examination, qRT-PCR and western blotting were utilized to inhibit MYC as a target of hsamiR-599. MYC expression was up-regulated in HCC, and introducing hsa-miR-599 could potentially reduce the mRNA and protein levels of the MYC gene. Additionally, over-expressed MYC could suppress the hsa-miR-599-induced effects on HCC-related properties in vitro. Overall, hsa-miR-599 acts as a TS and blocks HCC cell growth and migratory ability by partially targeting oncogenic MYC, which implies that hsa-miR599 can be a novel therapeutic option in the diagnosis as treatment of HCC [70].
Function of oncogenic miRs in hepatocellular carcinoma
Oncogenic miRs and their targeted genes in LC will help understand the effects of miRs in HCC as a diagnostic factor and potential targets for oncogenic miR-based management of HCC (Fig. 1, Table 1).
Function of miR-21
Meng et al. evaluated the expression profile of miRs in HCC and described the functional biological effect of miR-21 as a highly regulated miR and identified its target gene [71]. Using microarray tests in HCC tumors and cell lines and expression profile studies, they found that miR-21 had highly expressed miR in HCC [71]. Inhibition of miR-21 in HCC cells augmented the expression of PTEN and decreased the proliferation and migratory ability of tumor cells. In studies with induced expression of miR-21, which occurred by miR-21 mimic transfection, miR-21 increased tumorrelated properties. In addition, they found an increase in cell migration in normal human hepatocytes transfected with the miR-21 precursor. PTEN is a direct target of miR-21 and contributes to the effects of miR-21 on cell invasion. Modulation of miR-21 altered phosphorylation and expression of MMP-2 and MMP-9, both of which are downstream molecules of PTEN in migration and cell invasion. PTEN is involved in mediating the phenotypic features of cancer cells, such as cell growth and migratory ability, in aiding in the development and spread of HCC [71]. Also, miR-21 is the most naturally overexpressed miR in cancers. There are several mechanisms related to high miR-21 levels. The encoding genetic locus, 17q23, is strengthened in many tumors. In addition, miR-21 expression is produced by various cancer-related events, such as hypoxia and inflammatory responses. miR-21 is up-regulated in HCC cells and tissues, which are correlated with the migratory ability of HCC cells, where the miR-21 expression is inversely associated with the protein expression of its targeted genes and other signal molecules of its downstream cascades [72].
Function of miR-2
Hepatitis B virus (HBV)-miR-2 was upregulated in HCC patients with HBV infection and HBV-positive HCC cell lines [73]. Previous studies evaluated the oncogenic functions of miR-2 in cancer cells [74, 75]. To elucidate how HBV-miR-2 affects HCC progression, the effects of two target genes (TRIM35 and RAN) of HBV-miR-2 in LC cells were studied. The results showed that HBV-miR-2 promoted HCC cell growth by suppressing apoptosis and promoting migration and invasion through an improvement in the epithelial-mesenchymal transition (EMT), and acted as an oncogene in development of HBV-related HCC. Also, HBV-miR-2 suppressed the expression of TRIM35 but enhanced mRNA expression by targeting their 3′UTR and the ectopic expression of TRIM35 or knockdown of mRNA counteracted the malignant phenotypes induced by HBV-miR-2 [73].
Function of miR-221 and miR-222
Among oncogenic miRs that are up-regulated in LC, the tumor-induced activity of miR-221/mir-222 has been reported [76]. Also, miR-221 and miR-222 are two highly homologous miRs regulated in various human cancers, depending on the tumor system. miR-221 and miR-222 are known to influence the angiogenic functions of endothelial cells (ETCs) [77]. ETC migration and proliferation are controlled by miR-221 and miR-222, which regulate downstream targets. HCC cells overexpressed miR-221 and showed induction in cancer-associated properties. Previous studies investigated the association between enhanced expression of miR-222 and HCC progression, which acts through the AKT signaling induction [78, 79].
Function of miR-96
Iwai et al. analyzed the function of miR-96-5p in HCC [80]. It was noted that miR-96-5p expression was significantly up-regulated in HCC samples, which reached the rate of their adjuvant tissues. Gene amplification may be one of the primary mechanisms by which miR-96-5p expression can develop HCC. Transfection of miR-96-5p mimic into HCC cells reduced the CASP9 expression, which encoded caspase-9, the critical initiator caspase in the mitochondrial apoptotic cascade. A putative binding site for miR-96-5p was recognized in the CASP9 3’-UTR, and the luciferase assay showed that CASP9 is a possible target of miR-96-5p. The miR-96-5p mimics increased the resistance to doxorubicin (DOX) and ultraviolet-induced apoptosis by decreasing caspase-9 expression in HCC cells. Transfection of miR-96-5p inhibitor improved the cytotoxic effect of DOX by improving caspase-9 activity in the HCC cells, indicating a synergistic effect between the miR-96-5p suppressor and DOX. Therefore, miR-96-5p continually up-regulated in HCC inhibits apoptosis by targeting CASP9. Accordingly, miR-96-5p may be a possible therapeutic target for HCC [81]. Combined miR-96-5 p with AFP induced sensitivity of HCC detection. In addition, its serum levels were linked to tumor growth and migration. These results suggested that serum miR-96-5p can be used as a non-invasive biomarker for HCC detection [81].
Function of miR-17/92 cluster
The miR-17/92 cluster is known as oncomiR-1 due to its strong association with tumorigenesis. Six miRs from oncomiR-1 have been designated significant roles in numerous cellular activities via suppression of cell death in multiple cancer-related processes [82]. Li et al. reported that triptolide inhibited cell proliferation and induced significant apoptosis in multiple HCC cells with various p53 statuses. Triptolide treatment modified numerous targets implicated in other cascades. In an animal model, there was reduced tumor size in the triptolide-treated group compared to the controls. Two miR clusters, miR-17/92 and miR-106b/25, were inhibited considerably by triptolide, which resulted in up-regulation of their normal target genes, including BIM, PTEN, and p21. In the LC sample, increased levels of these miR groups are associated with shorter recurrence-free survival. Triptolide suppressed the expression of these miRs in a c-MYC-dependent fashion, which improved triptolide-induced cell death. Also, triptolide down-regulated the expression of c-MYC by targeting other intermediating proteins [83].
Function of miR-27
Aberrant expression of miRs affects HCC development and progression. miR-27b was discovered previously to play essential roles in human tumors. Nevertheless, its expression level, clinical effectiveness, and biological processes in HCC are still unclear. The expression and cancer-related properties of miR-27b in HCC specimens and cells were assessed using qRT-PCR, MTT, and BrdU proliferation assays. An animal model was used to evaluate the HCC tumor growth in vivo. TargetScan and a luciferase reporter assay revealed the putative target gene of miR-27b. miR-27b was overexpressed in HCC. Overexpression of miR-27b was associated with adverse prognostic characteristics and a reduction in survival rate. Inhibiting miR-27b in SMMC-7721 cells significantly repressed proliferative ability and cell-cycle progression while improving apoptosis. In contrast, miR-27b overexpression improved the proliferation and function of the cell cycle and reduced apoptosis of Hep3B cells. In xenograft animal models of SMMC-7721 inoculated cells, miR-27b suppression caused inhibition of tumor growth. Also, Fbxw7 was regulated by miR-27b in HCC. It has been suggested that miR-27b can serve as an oncogenic miR in HCC by modulating cancer-associated properties, and its downstream target gene, Fbxw7, mediates its oncogenic effect [84]. The importance of miR-27b in regulating liver fat metabolism, both in healthy and cancer tissue, has been demonstrated; however, the mechanisms by which this modulation of the lipid profile of cells is performed are not yet well understood. The effective translocation of miR-27b has been identified by noting a significant decrease in the EGFR expression of the target miR-27b gene, and a significant increase in the total amount of available triglycerides, with a trend toward saturation, was observed [85]. No significant changes were observed in any other fat groups, and it has been shown that triglyceride metabolism is the primary target of regulation by miR-27b, which warrants further investigation [85].
Function of miR-490
miR4903p has been identified as an oncogene or TS in different kinds of cancer, including LC. In some studies, low expression levels of both miR-490-3p and miR-490-5p were observed in HCC [86]. The TS and oncogenic mechanism of miR-490 involves the regulation of oncogene and TS gene expression. miR4903p has a TS and promoter in a context-dependent manner in HCC [87]. Bioinformatic studies analyzed the affinity between PPM1F and miR-490-3p expression. Other bioinformatic data evidenced that circSLC3A2 binds to miR-490-3p. The dual-luciferase assay and RNA immunoprecipitation approaches proved the binding sites among miR490-3p and PPM1F or circSLC3A2. The localization and clinical importance of miR-490-3p and circSLC3A2 in HCC patients were examined by fluorescence in situ hybridization. MTT, soft-agar, and Transwell assays were carried out to assess the effects of miR-490-3p or circSLC3A2 on the migratory ability of the cells. miR-490-3p reprogramming by targeting PPM1F prevented HCC-related activity, but its blocker inhibited these functions. In addition, circSLC3A2, predominantly localized in the cytoplasm, showed an oncogenic effect by absorbing miR-490-3p, regulating PPM1F expression, and reversing the HCC-associated properties [88].
Function of miR-494
Research shows that miR-494 is overexpressed in human HCC and is associated with regulating G1/S cell cycle transmission by targeting mutant TS in colorectal cancer. Inhibiting miR-494 in human HCC cell lines reduces cell cycle progression, and anti-miR-494 treatment significantly reduces primary tumor formation led by MYC and this miR was reported as a new therapy option [89]. Also, miR-494 was identified as an oncogenic miR in LC, mediating the side effects of its migration, mainly by reducing an enzyme that regulates epigenetic labeling, followed by inhibition of several invasive suppressor miRs. Thus, disruption of miR-494 regulation may act as an initial event for a cascade leading to HCC cell development. Based on miR profiles of HCC patients with and without vascular invasion as well as data derived from HCC cell lines, interestingly, overexpression of miR-494 increased the methylation of proximal islets of CpG and consequently decreased miRs, which are known suppressors of cell invasion. miR-494 may affect these miRs by inhibiting promoter demyelination. Using bioinformatics tools, the TET family of methylcytosine dioxygenases was identified as the potential target of miR-494. This family of enzymes converts 5-methyl cytosine (5mC) to 5’-hydroxymethyl cytosine (5mC) and removes the epigenetic label [90].
Function of miR-675
Earlier growth response-1 (EGR1) is a multi-domain protein and an immediate-early transcription factor induced during liver injury and controls the expression of various genes implicated in metabolism, cell proliferation, and tumorigenesis. An innovative study focused on the function of EGR1 in LC development and the underlying mechanism of action. Two LC-related datasets, GSE101728 and GSE138178, obtained from the Gene Expression Omnibus (GEO) database, were used to specify essential genes involved in cancer progression. Microarray analysis was conducted to determine differentially expressed miRs after EGR1 knockdown [91]. The target gene of miR-675 was identified by integrated analysis. EGR1 and miR-675 were highly expressed, whereas sestrin 3 (SESN3) was poorly represented in LC tissues and cells. High EGR1 expression was associated with poor liver function and disease severity in patients with LC. Knockdown of EGR1 weakened the proliferation and invasiveness of LC cells. EGR1 binds to the miR-675 promoter and increases its transcription, and miR-675 binds to SESN3 mRNA to induce its down-regulation. Also, miR-675 up-regulation promoted the malignancy of LC cells, but further up-regulation of SESN3 reduced the invasiveness of cells. SESN3 was enriched in Wnt/β-catenin signaling. EGR1 and miR-675 activated Wnt/β-catenin through down-regulating SESN3. This analysis demonstrated that EGR1 promotes the malignant behaviors of LC cells by mediating the miR-675/SESN3/Wnt/β-catenin axis [91].
Discussion
The role of tumor immunity in HCC pathogenesis and the well-characterized function of immune-suppressive cells such as regulatory T cells that are crucially implicated in the pathogenesis of HCC, lead to a major shift in HCC development, progression, and treatment [92]. Future HCC trials based on targeting regulatory T cells by new immunotherapies based on immune checkpoint inhibitors (ICI) with fewer immune-related adverse events are needed for successful therapy of HCC. Granito et al. highlighted the current clinical trials based on ICI agents as treatment for HCC [92]. Also, Stefanini et al. recently described and summarized the enhanced anti-tumor effects of combination treatment based on tyrosine kinase inhibitors (TKIs) plus ICIs obtained in recent and ongoing clinical trials [93]. TKIs such as sorafenib were the first approved systemic treatments for the advanced stage of HCC; however, other systemic treatment of HCC has been further expanded with a combination of TKIs with ICI, which make them a good candidate for HCC combination therapy. Current and future studies on miRs could improve current treatment strategies based on immunotherapy as well as combination treatment strategy based on ICIs plus TKIs/anti-angiogenic agents. In the future, more well-designed research focusing on the translational potential of miRs in HCC treatment will be required.
Angiogenesis and metastasis play a pivotal role in HCC development and recurrence. miRs are abundant in ETCs, and several studies have revealed that they regulate vascular functions in the tumor microenvironment. Numerous miRs have been demonstrated to have proangiogenic or antiangiogenic properties. The miR-15a/16-1 cluster can induce apoptosis by targeting Bcl-2, cyclin D1, and some other signaling pathways; although certain miRs inhibit angiogenesis, others encourage the growth of new blood vessels. Moreover, miR-126, as endothelium-explicit miR, can control VEGF expression levels, and ETC expression while downregulates tumor vascularization and neoangiogenesis. miR-296 can change the outflow of VEGF receptor 2 and platelet-derived growth factor receptor by directly focusing on the HGF-managed tyrosine kinase substrate. The first cancer-causing miR was identified in the miR-17/92 cluster, which includes miR-17, miR-18, miR-19a, miR-19b-1, 20a, and miR-92-1. Although the significant function of the miR-17/92 group is to direct the transcription factors c-MYC and E2F and their autoregulatory circle, it likewise advances growth angiogenesis by focusing on thrombospondin-1, connective tissue development factor, and numerous other proangiogenic targets.
Many miRs have already been investigated in HCC [94]. Some miRs have been linked to metastasis via the modulation of EMT. Five members of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429), as well as miR-205, can induce E-cadherin expression, block EMT, and restrict cancer cell motility and invasion. However, miR-155 increases cancer cell expression by targeting the Ras homolog gene family member A and plays a role in tumor growth factor (TGF)-induced EMT, cell motility, and metastasis [95]. In opposition to this miR, miR-124 inhibits cytoskeleton rearrangement and EMT via inhibiting ROCK2 and EZH2, reducing HCC’s invasive and metastatic potential [96]. miR-29a can increase EMT and cancer cell migration in coordination with oncogenic Ras signaling mediators [97].
miRs, which regulate cell growth and apoptosis, might play a pivotal role in cancer cell survival and arrest in the bloodstream throughout the metastatic process. miR-126 reduces leukocyte and cancer cell attachment to ETCs by targeting adhesion protein VCAM-1, which is typically down-regulated in malignancies. Numerous miRs have also been shown to play important roles in T cell and B cell activation, as well as innate and adaptive immune responses. In both conventional and regulatory T cells, miR-155 is required for T lymphocyte functions, B lymphocyte production, and TLR responsiveness. Cancer-suppressing miR-146a, which controls TLR and cytokine signaling, inhibits TRAF6 and IRAK1 genes [98]. Essential immune system regulators miR-150, miR-17/92 clusters, and miR-181 may have a role in cell survival and migration [99, 100].
Accumulated evidence was discussed in terms of the current challenges and feasibility of using miRs as therapeutic agents in HCC [101]. There are numerous findings regarding pre-clinical experiments; however, the data for examining the efficacy of miR-based treatment of HCC is limited. A major obstacle is the delivery system for miR transfection and its translation into clinical settings [102, 103]. This miR-based strategy is still at an early stage of development, and there have been some miR-based clinical trials for HCC therapy [103], but no miR-based therapies have been proven for clinical treatment of HCC to date.
Conclusions
In this review, we addressed the current knowledge of the role of miRs in HCC development and progression. The accumulated evidence has been considered as regards using miR-based strategies for HCC diagnosis and therapy. Despite advances in this field of study, some issues are still unsolved. It is vital to ensure that miR delivery systems be both effective and safe for people. Because miRs are involved in the pathogenesis of HCC, researchers should consider the establishment of regulatory networks in liver tumors that comprise genes and miRs. This would allow for a better understanding of cancer’s molecular properties. As a result, in the future, more well-designed research focusing on translational potential will be required. Removal of technical variations can improve miR identification accuracy. In addition, further large randomized prospective clinical trials are needed to assess the potential value of miRs in HCC treatment, particularly for HCC gene therapy. However, the miR research is confronted with problems. Many studies are still in the experimental stage and will not lead to application in clinical practice. Second, more studies are needed regarding the security and reliability of miRs as therapeutic indicators for HCC and early detection of HCC. Furthermore, while most research on miR content in serum exosomes has focused on exosomal miRs, their specific mechanisms of HCC remain unclear. The lack of sensitive exosome preparation and analysis methods further impedes clinical translation. As a result, future research should concentrate on converting miR discoveries in HCC into practical applications, such as creating therapeutically available miR inhibitors. Furthermore, priority should be given to studying the security and reliability of miRs for early HCC detection and treatment. Future studies must consider exosome-mediated miR transmission and improvement in analytical technologies.






