Introduction
Rheumatoid arthritis (RA) is described as a common autoimmune disease, characterized by chronic, symmetric joint inflammation. It causes bone and cartilage destruction leading to functional disability and is considered as an individual, social and economic problem [1, 2]. RA occurs in 1% of the population worldwide with female predominance [3]. The risk of RA can increase up to 60% in people with certain genetic factors [4].
One of the most significant genetic links to the disease is the HLA-DRB1 shared epitope and the major hypothesis describing it was enunciated in the late 1980s. The shared epitope (SE) is a specific five amino acid sequence in positions 70 to 74 within the antigen-binding groove of HLA-DR-b chains [5].
The shared epitope is responsible for antigen presentation, T-cell repertoire and self-peptide selection and may have a role in autoreactive adaptive immune response stimulation. HLA-DRB1 is recognized as being associated with RA in those patients who are positive for the rheumatoid factor (RF) or anti-cyclic citrullinated peptide (anti-CCP) [6]. It was found that 70% of anti-CCP positive patients possessed one of the SE variants [7]. It seems that anti-CCP positive and anti-CCP negative disease types are different in terms of the genetic background of RA development [8].
Furthermore, the strength of association is different for separate allelic variants. In North Europeans, a powerful risk variant is HLA-DRB1*04:01, especially within the HLA-DRB1*04:04/04:01 heterozygous genotype. In contrast, DRB1*13:01 seems to play a protective role in anti-CCP positive disorder [9, 10]. Among Greek RA patients, DRB1*04:05, DRB1*01:01 and DRB1*10:01 belong to the most frequent SE alleles [11]. This is not surprising, as in general there are differences in HLA-DRB1 allele distribution between the Polish and southern European populations [12].
Despite that, environmental factors such as smoking, which has been identified as the main inductor of citrullination, may be harmful in combination with HLA-DRB1 molecules and promote the autoimmune reaction [13].
Variability within the KLRK1 gene coding for an activator of the immune system — Natural Killer Group 2, Member D (NKG2D), a receptor belonging to the C-type lectin-like family of transmembrane proteins – constitutes another RA related genetic factor. The NKG2D receptor is expressed on the natural killer (NK) cell surface as well as CD8+T cells and γδ+ T cells [14]. The T lymphocytes and NK cells, both of which express NKG2D, are recognized as being an important part of RA pathogenesis [15]. This may suggest potential involvement of NKG2D in RA. Increased numbers of unique CD4+ cells lack the CD28 molecule and express the NKG2D receptor. It was also observed in peripheral blood and synovial tissue of patients suffering from this disease [15, 16].
NKG2D is associated with multiple ligands, but among the first described were MHC class I-chain related proteins, MICA and MICB, with the ability to activate NK cells [17]. Exposure to cellular stress, infection, inflammation and DNA damage upregulate MIC expression [18]. Previously investigated single nucleotide polymorphisms in MICA and NKG2D regions suggested an association with susceptibility to RA in German and Indian patients [19, 20]. Nonetheless, to the best of our knowledge, there has been no study examining MICA and NKG2D variants in both Polish and Greek populations.
Our recent findings regarding the effect of MICA [21] and NKG2D polymorphism on RA treatment outcome [22] have prompted us to analyse and compare the genetic variability of Greek and Polish patients with respect to the presence of the HLA-DRB1 SE as well as MICA and NKG2D variants.
Material and methods
Patients and controls
61 Greek and 100 Polish RA patients, diagnosed according to the 2010 American College of Rheumatology (ACR)/European League Against Rheumatism (EULAR) criteria, were investigated. Collected data of the patients are summarized in Table 1. The study was approved by the Wrocław Medical University Ethics Committee and Evangelismos Hospital.
Table 1
Characteristics of Polish and Greek rheumatoid arthritis (RA) patients
Genotyping
The single nucleotide polymorphisms (SNPs) in the genes coding for MICA and its receptor, NKG2D, were selected based on available literature analysis as well as search results from the NCBI Database of Short Genetic Variations (dbSNP). Information regarding predicted functional consequences of SNPs was obtained using the SNPinfo Web Server [23]. The following SNPs were selected: MICA rs1051792 (G>A; missense variant in exon 3 – Val129Met), KLRK1 rs1154831 (C>A; intronic variant; potential transcription factor binding site), KLRC4-KLRK1 rs1049174 (C>G; 3' untranslated region (UTR); potential miRNA binding site) and KLRC4-KLRK1 rs2255336 (A>G; nonsynonymous polymorphism in exon 4 – Thr72Ala).
Genotyping for MICA rs1051792, KLRK1 rs1154831 and KLRC4-KLRK1 rs1049174 was performed using a LightSNiP assay (TIB MOLBIOL, Germany). The KLRC4-KLRK1 rs2255336 was studied employing a TaqMan SNP genotyping assay (Thermo Fisher Scientific, Waltham, MA, USA). The LightCycler 480 Real-Time PCR system (Roche Diagnostics, Rotkreuz, Switzerland) was used in this analysis. HLA-DRB1 alleles were genotyped by polymerase chain reaction (PCR) followed by hybridization with sequence-specific oligonucleotide probes (PCR-SSOP), PCR with sequence specific primers (PCR-SSP) or sequence-based typing (SBT). Genotyping results were correlated with clinical parameters.
Statistical analysis
Potential associations between examined SNPs and clinical parameters of RA patients were analysed by Fisher’s exact test for parametric values. The same test was also used to compare genotype variation distribution within patients. P-values less than 0.05 were considered statistically significant. All statistical calculations were performed using the GraphPad7 Prism software.
Results and discussion
HLA-DRB1 SE has been of particular interest for many years, especially in regard to its association with RA [10, 11].
Table 2 depicts SE risk alleles in both Greek and Polish RA patients. As expected, the majority of patients, 62.3% of Greek and 72.0% of Polish RA cases of the present study, were characterized by the presence of HLA-DRB1 SE alleles. Also in line with the results of the previous studies, involving both studied populations of RA patients, DRB1*01:01, *04:05 and *10:01 were predominantly represented among Greek patients [24-26] while DRB1*01:01 and *04:01 alleles were predominantly represented among Polish patients [10]. The latter DRB1*04:01 allele was associated with RA susceptibility in various populations as shown, for example, also for UK patients [27]. According to another study in UK patients, HLA-DRB1*04:04 is more frequently observed than HLA-DRB1*01:01, especially in anti-CCP-positive and RF-positive RA patients [28]. On the other hand, in Korean RA patients, HLA-DRB1*04:05 correlated with the disease [29], whereas other HLA-DRB1 alleles, such as *04:05, *04:01, *09:01, *01:01, *14:01, *16:02, *04:03, and *14:05, were significantly associated with RA in the Japanese [30]. These results underline the differences in HLA-DRB1 SE segregation between various populations and confirm the predominant representation of specific HLA-DRB1 SE alleles associated with the RA risk among Greek and Polish patients.
Table 2
HLA-DRB1 shared epitope frequency in rheumatoid arthritis (RA) patients
For the past few years, non-classical HLA polymorphism, including MICA and NKG2D, has been investigated in Polish patients with RA [21, 22, 31]. As those publications were focused only on one population, there is still a lack of knowledge about the effect of these genetic factors in Greeks [11, 26]. Therefore, in the present study, MICA and NKG2D polymorphisms were also analysed in a group of Greek patients.
Firstly, we compared the distributions of MICA alleles and their association with HLA-DRB1 alleles in both groups of patients. As shown in Table 3, no significant differences were observed in genotype distribution between Polish and Greek patients within MICA rs1051792. In both groups the AA homozygous MICA genotype was the rarest.
Table 3
Distribution of MICA and NKG2D alleles, genotypes in rheumatoid arthritis (RA) patients
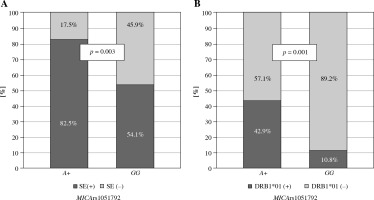
No significant differences were observed in the presence of the SE among Greek patients with various MICA rs1051792 alleles or genotypes. However, the Polish A allele carriers were more likely to possess one of the HLA-DRB1 SE alleles (p = 0.003), particularly DRB1*01:01 and/or DRB*01:02 (p = 0.001) (Fig. 1).
Fig. 1
Relationship between MICA rs1051792 genotype and presence of HLA-DRB1 SE alleles in Polish RA patients. MICA A allele carriers were more likely to possess one of the HLA-DRB1 SE alleles, (A) especially HLA-DRB1*01:01 and/or DRB1*01:02 (B)
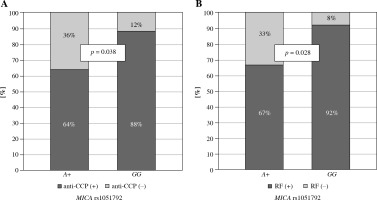
Secondly, MICA rs1051792 polymorphism was investigated for a potential association with clinical parameters in both Polish and Greek RA patients. A subsequent analysis revealed that the GG homozygotes are more frequent among Greek patients with anti-CCP antibodies and RF as compared to those carrying the A variant (p = 0.038 and p = 0.028, respectively) (Fig. 2). It has been also documented by Alexiou et al. that a higher level of anti-CCP antibodies may have prognostic significance in Greek patients with extra-articular manifestation [32] as well as radiographic joint damage [33].
Fig. 2
Relationship between MICA rs1051792 genotype and clinical parameters in Greek RA patients. MICA GG homozygous patients more frequently presented with anti-CCP antibodies (A) and RF (B) as compared to those carrying the A variant
On the other hand, these relationships between anti-CCP and RF positivity and MICA GG homozygosity detected in Greeks were not seen in the Polish group of RA patients. Note, however, the results of our previous study by Iwaszko et al. documenting that MICA rs1051792 polymorphism can affect the efficiency of anti-tumour necrosis factor (TNF) therapy in Polish patients [21]. Thus this MICA polymorphism seems to play a significant role also in Polish RA patients. In a German and French cohort, the MICA rs1051792 G allele was also associated with disease development risk [19]. Interestingly, the correlation between GG homozygotes and RF presence was shown in Tunisians [34], which is in line with our results for the Greek cohort.
In the third part of the study, we focused on analysis of the NKG2D genetic variants. The mechanism and functions of MICA-128Met/Val (G/A) dimorphism in disease pathogenesis remain partly obscure [35]. However, it has been documented that the protein encoded by the MICA-129-Met (A) allele is characterized by stronger interaction with NKG2D than the alternative Val (G) isoform. Therefore, downregulation of NKG2D by the Met (A) variant is more efficient.
The role of NKG2D genetic variants, including those selected in the present study (rs1049174, rs2255336), has been previously investigated by Mariaselvam et al. in regard to RA susceptibility in an Indian population. However, this study did not reveal any statistically significant correlation between those single nucleotide polymorphisms and clinical parameters of the disease [20].
In this present study, the NKG2D rs1049174 G variant and GG homozygosity were more frequent among the Greeks than the Polish patients (p < 0.001) (Table 3). However, no significant differences were observed in genotype distribution between Polish and Greek patients within NKG2D rs1154831 and NKG2D rs2255336.
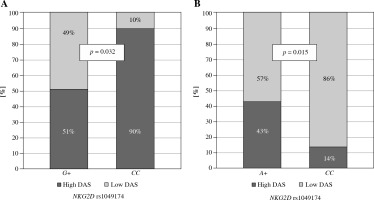
Moreover, the NKG2D rs1049174 G variant was found to be associated with lower disease activity score in Greek RA patients (p = 0.032, Fig. 3A). In addition, NKG2D rs1154831 CC homozygotes were also characterized by a lower disease activity score (p = 0.015, Fig. 3B). This is a novel observation not previously described. This finding is supported by a Korean study conducted by Park et al., where presence of NKG2D rs2255336 wild type homozygosity, but not NKG2D rs1049174, resulted in an increased risk of RA [36].
Fig. 3
Relationship between disease activity score and NKG2D genotypes in Greek RA patients. Lower disease activity score characterised patients carrying the NKG2D rs1049174 G variant (A) or being NKG2D rs1154831 CC homozygotes (B)
Summarizing the above, HLA-DRB1 SE alleles are well-documented risk factors for RA development, but the association of particular alleles varies between populations. The vast majority of Polish and Greek patients with RA are positive for SE alleles, but DRB1*01:01, *04:05 and *10:01 predominate among Greek while DRB1*01:01 and *04:01 predominate among Polish patients. Differences between the two populations are also seen with respect to the other genetic loci. In Polish patients the presence of DRB1 SE is associated with the MICA rs1051792 A allele. Among Greek patients, the NKG2D rs1051792-GG homozygotes more frequently presented with anti-CCP antibodies and RF, while carriers of the NKG2D rs1049174-G variant and rs1154831-CC homozygotes were characterized by lower disease activity scores (p < 0.05 in all cases).
In conclusion, the results of the present study show that HLA-DRB1 SE alleles, as well as MICA and NKG2D genetic variants, are associated with RA development and their frequencies differ between the Greek and Polish populations of RA patients.


