Introduction
With over 1.1 million affected children and adolescents worldwide (IDF Diabetes Atlas Eight Edition 2017), type 1 diabetes (T1D) is one of the major chronic disorders of the youth. The condition is caused by an autoimmune destruction of the pancreatic β cells. It requires life-long substitution with exogenous insulin and leads to serious complications that usually progress over the course of disease. The etiology of T1D remains unclear, and results from interplay between complex genetic predisposition and poorly defined environmental triggers, which probably misguide the immune system. Human leukocyte antigen (HLA) alleles account for 40-50% of the genetic risk to develop T1D, and the majority of other known susceptibility loci also harbor genes involved in the immune response and lymphocyte function in particular [1, 2].
Signal transducer and activator of transcription 4 (STAT4) gene maps to chromosome 2q32.2-32.3 and encodes a member of the family of latent cytoplasmic transcription factors, which become phosphorylated in response to pro-inflammatory cytokines. Once activated by the interleukin (IL)-12, key molecule of the cell-mediated immune response, phosphorylated STAT4 homodimerizes, translocates to the nucleus and interacts with specific DNA sequences to modulate target gene transcription [3]. STAT4 primarily promotes Th1 cell differentiation and interferon (IFN)-γ production that is critical for most inflammatory reactions [4, 5]. It also plays a role in the development of Th17 cells, probably through its implication in IL-17 and IL-23 signaling, which seems essential for autoimmunity-associated inflammation, notably in T1D [6, 7]. STAT4-deficient mice present defective IL-12-mediated IFN-γ induction, decreased NK cell cytotoxicity, and impaired Th1 cell expansion [8]. Since both IL-12 and Th1 cells are involved in the initiation and acceleration of diabetes in NOD mice model, STAT4 emerges as a likely functional candidate for autoimmune diabetes [9, 10]. In accordance, the genome-wide transcriptome analysis of CD4+ T cells from the NOD mice identified the expression of IL-12 and STAT4 at 4 weeks of age, prior to the development of insulitis [11]. On the contrary, STAT4-depleted NOD mice or STAT–/– mouse model of virus-inducible diabetes demonstrate significantly reduced production of IFN-γ, decreased or absent insulitis, and preserved insulin secretion [12, 13].
Therefore, altered STAT4 expression may be implicated in disturbed immune function, and contribute to autoimmunity. In humans, STAT4 is expressed in several tissues including spleen, brain, testes, and peripheral blood mononuclear cells (PBMCs). Its level is low in resting lymphocytes and only rises upon stimulation, in parallel with the up-regulation of the high-affinity IL-12 receptor [3, 14]. Polymorphisms in STAT4 gene may exert an impact on its expression and hence affect the immune function [15-17]. In line, genetic variants of STAT4 are associated with variety of autoimmune disorders, including systemic lupus erythematosus (SLE) [15, 18], rheumatoid arthritis [18-20], systemic sclerosis [21], and Sjögren syndrome [22]. Organ-specific autoimmune conditions have received much less attention with this regard, although STAT4 associations have been reported for celiac disease [23], autoimmune thyroid disease [24, 25], and Addison’s disease [26]. Most of these associations pointed out rs7574865 as a tagging STAT4 single nucleotide polymorphism (SNP) in autoimmunity. The analyses performed in T1D provided inconsistent results. Although STAT4 locus has not been picked up in the genome-wide association studies, it displayed association with T1D in some populations. This was further supported by the results of meta-analyses, but considerable population differences were still described [20, 27-33]. To the best of our knowledge, no data are available from the Central European cohorts of diabetic patients to date.
The aim of this study was to investigate the association of STAT4 with T1D by genotyping of rs7574865 and through gene expression analysis in Polish children with newly diagnosed disease. Furthermore, considering that STAT4 polymorphism seemed to be a risk factor for several autoimmune disorders, an attempt was made to evaluate STAT4 variant and gene expression with regard to co-occurring thyroid autoimmunity in T1D patients. Autoimmune thyroid disease is the most frequent concomitant condition, with serum anti-thyroid antibodies detectable in up to 18% of children and adolescents with T1D [34, 35]. Thyroid autoimmunity and autoimmune diabetes share several genetic susceptibility loci [36].
Material and methods
Genotyping of rs7574865 was performed in 656 children and adolescents with T1D (345 females and 311 males), and in 782 healthy adult controls (419 females and 363 males) with no history of diabetes. All study participants were Polish of Caucasian ethnicity. The diagnosis of T1D was based upon the WHO criteria with absolute dependence on exogenous insulin confirmed. Mean age (±SD) at T1D onset was 8.2 ±4.2 years, and mean age at the time of the study was 11.4 ±5.5 years. Patients were enrolled between 2010 and 2016 at the Department of Pediatric Diabetes and Obesity, Poznan University of Medical Sciences, Poland. Control subjects were recruited among healthy blood donors at the regional blood transfusion center in Poznan. Healthy controls presented negative history of autoimmunity, no signs or symptoms of the autoimmune or chronic inflammatory disease, and normal fasting plasma glucose at the time of the study.
Subsequent expression analysis was conducted in 29 children with newly diagnosed T1D (mean age ±SD: 11.2 ±3.3 years) and 27 age-matched healthy controls (mean age, ±SD: 10.5 ±4.1 years). STAT4 mRNA expression was investigated in their PBMCs separated from the peripheral blood by density gradient centrifugation on Histopaque-1077 (Sigma Aldrich, Germany). To minimize the influence of the acute metabolic imbalance at T1D diagnosis, the blood was collected from fully rehydrated patients with normalized ketonemia and well-controlled glycaemia, 14 days after the introduction of insulin therapy, when the autoimmune process was still ongoing. Samples from the age-matched children with negative personal and family history of autoimmunity, no clinical signs of autoimmune disorders, and fasting plasma glucose within the reference range were obtained from the local outpatient pediatric practice. Only subjects with no clinical symptoms of infection and negative inflammatory tests (C-reactive protein, complete blood count) were included in the study and control group.
The Ethics Committee at the Poznan University of Medical Sciences approved the study protocol and all study procedures remained in accordance with the Declaration of Helsinki. Informed consent was obtained from the participants aged at least 16-years-old and, in case of minors, from their legal representatives.
Serum autoantibody evaluation
Serum samples from T1D patients diagnosed between 2013 and 2016 were analyzed using the same set of validated, commercially available radioimmunoassays. β cell-specific autoantibodies were assessed using radioimmunoassays (RIA) to detect antibodies against insulin (IAA), glutamic acid decarboxylase (GAD), and islet antigen 2 (IA-2). IAA were analyzed with IAA RIA kit from DIAsource Immuno Assays (Louvain-la-Neuve, Belgium), whereas GAD and IA-2 with Anti-GAD and Anti-IA2 RIAs from Euroimmun (Lubeck, Germany). RIA technique was also used to test for markers of thyroid autoimmunity. Antibodies to thyroperoxydase (aTPO) and thyroglobulin (aTg) were evaluated by means of BRAHMS anti-TPOn and anti-Tgn RIA assays (Berlin, Germany), respectively.
Genotyping
Genomic DNA was extracted from the peripheral blood using Gentra Puregene Blood Kit (Qiagen, Hilden, Germany). Genotyping of rs7574865 was performed by real-time PCR on CFX96 real-time detection system (BioRad Laboratories, CA, USA), using validated TaqMan SNP genotyping assay (C_29882391_10), and following the conditions recommended by the manufacturer (Applied Biosystems by Thermo Fisher Scientific, USA). Genotypes were confirmed by direct DNA sequencing of both strands by BigDye terminator cycle sequencing ready reaction kit on ABI Prism 3730 genetic analyzer (Foster City, CA, USA). To ensure the accuracy of genotyping, 7% of samples were re-genotyped blind.
Gene expression analysis
The expression of STAT4 mRNA in PBMCs was evaluated by quantitative reverse-transcription PCR (qRT-PCR). Total RNA was extracted using QIAamp RNA blood mini kit (Qiagen, Germany). The quantity and the quality of RNA were estimated by spectrophotometric measurements (NanoDrop, Thermo Scientific, USA) and agarose gel electrophoresis, respectively. RNA was then converted to cDNA with QuantiTect reverse transcription kit (Qiagen, Germany).
qRT-PCR was performed using 10 ng of cDNA, Master Mix HOT FIREPol Solis 5x (Solis Biodyne, Tartu, Estonia)and TaqMan gene expression assay (Hs01028017_m1, Applied Biosystems by Thermo Fisher Scientific, USA) in a total volume of 20 µl. PCR reactions were run in triplicate on BioRad CFX96 real-time PCR instrument. The thermal cycling conditions comprised initial uracil-N-glycosylase activation for 2 minutes at 50°C, polymerase activation for 20 seconds at 95°C, followed by 40 cycles of 3 seconds at 95°C and 30 seconds at 60°C. β-2-microglobulin (B2M) mRNA (assay Hs00187842_m1) was quantified as control to normalize for the differences in mRNA levels. Mean cycle threshold (Ct) values were estimated with BioRad CFX Manager 3.1 software. Relative expression levels were calculated using the 2-ΔCt formula [37].
Statistical analysis
The sample size calculation, performed with PS power and sample size calculator v.2.1.30 (Vanderbilt University, TN) assuming an allelic OR of 1.3 reported in the former studies [20, 31] and 80% power to detect that effect, showed minimal sample size of 618 cases and 737 controls (p = 0.05). Genotypes were checked for Hardy-Weinberg equilibrium (threshold p> 0.05) using an online calculator available at the Helmholtz Center Munich website (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl).
Statistical analyses were performed using GraphPad Prism 6.0c (GraphPad Software, La Jolla, CA). χ2 test was used for association analysis on 2 × 2 and 2 × 3 contingency tables. In the case of low cell counts, Fisher exact test was applied, with Freeman-Halton extension to compute the probability of obtaining a distribution of values in a 2 × 3 contingency table. Normality of continuous data was checked with Shapiro-Wilk test. Normally distributed data were further compared using Student’s t-test for unpaired samples, whereas those with non-normal distribution were analyzed by non-parametric Mann-Whitney test. Genotype-stratified normally distributed data were compared using one-way ANOVA, and those with non-normal distribution were analyzed by the Kruskal-Wallis test. Two-tailed p values < 0.050 were considered statistically significant.
Results
Complete data of the three diabetes-specific autoantibodies evaluated with the same set of assays were available from 374 individuals with T1D. Positive IAA were found in 190 (50.8%) affected children, while GAD – in 243 (65.0%) and IA2 – in 262 (70.1%) T1D patients, respectively. All three studied autoantibodies were positive in 75 (20.1%) patients, whereas two circulating antibody specificities were found in 171 (45.8%) individuals at disease onset. Positive markers of thyroid autoimmunity, aTPO and aTg, were detected in 16.9% and 11.4% of T1D patients, respectively, with both thyroid antibodies found in 9.5% subjects.
STAT4 rs7574865 genotypes in both studied cohorts conformed to the Hardy-Weinberg equilibrium (p = 0.351 for T1D group and p = 0.774 for controls). The distribution of rs7574865 genotypes among patients vs. controls demonstrated significant difference (p = 0.002). In line, rs7574865 minor T allele was found in 25.2% T1D patient alleles compared to its 19.5% prevalence among healthy subjects (OR, 1.39 [95% CI: 1.16-1.66], p< 0.001). Detailed genotype and allele data are displayed in Table 1. Moreover, carriers of the rs7574865 minor allele presented an earlier T1D onset compared to the wild type homozygotes (mean age: 6.8 ±3.9 years for TT, 8.1 ±4.3 years for GT, and 8.5 ±4.1 years for GG subjects; p = 0.017). On the contrary, no significant differences were found in genotype-stratified patients with regard to their GAD(p = 0.991), IA-2 (p = 0.820), and IAA (p = 0.223) autoantibody status at the clinical beginning of the disease. Furthermore, no differences in rs7574865 frequencies were detected in individuals with one, two, or three positive β cell autoantibodies upon T1D diagnosis (p = 0.516). However, both aTPO and aTg antibodies were significantly more frequent in carriers of the minor T allele (p = 0.039 and p = 0.007, respectively) (Table 2).
Table 1
Distribution of alleles and genotypes of the STAT4 gene polymorphism in 656 patients with type 1 diabetes (T1D) compared to 782 healthy controls (CON)
| rs7574865 | T1D (%) | CON (%) | p-value |
|---|---|---|---|
| GG | 372 (56.7) | 508 (65.0) | 0.002 |
| GT | 238 (36.3) | 243 (31.1) | |
| TT | 46 (7.0) | 31 (3.9) | |
| G | 982 (74.8) | 1259 (80.5) | < 0.001 |
| T | 330 (25.2) | 305 (19.5) |
Table 2
Distribution of genotypes and alleles of the STAT4 gene polymorphism in patients with type 1 diabetes and thyroid-specific autoantibodies against thyroperoxidase (aTPO) and thyroglobulin (aTg)
| rs7574865 STAT4 | aTPO (n = 331) | aTg (n = 317) | |||
|---|---|---|---|---|---|
| aTPO-positive 56 (%) aTPO-negative 275 (%) | aTg-positive 36 (%) | aTg-negative 281 (%) | |||
| GG | 28 (50.0) | 175 (63.6) | 14 (38.9) | 178 (63.3) | |
| GT | 21 (37.5) | 87 (31.6) | 17 (47.2) | 89 (31.7) | |
| TT | 7 (12.5) | 13 (4.7) | 5 (1.9) | 14 (5.0) | |
| p-value | 0.039 a | 0.007 a | |||
| G | 77 (68.8) | 437 (79.5) | 45 (62.5) | 445 (79.2) | |
| T | 35 (31.2) | 113 (20.5) | 27 (37.5) | 117 (20.8) | |
| p-value | 0.013 b | 0.002 b | |||
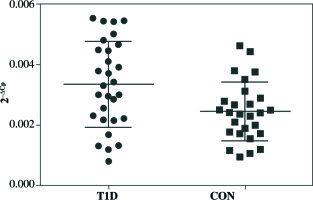
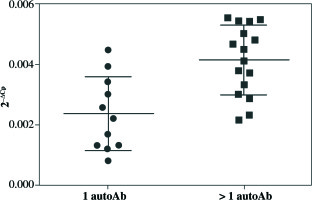
qRT-PCR analysis revealed significant up-regulation of STAT4 mRNA in T1D patients’ PBMCs compared to healthy controls (fold change 1.37, p = 0.008 when normalized for the B2M expression level) (Fig. 1). No differences in STAT4 expression were found in patients positive for serum GAD, IA-2 or insulin autoantibodies; however, those who displayed 2 or 3 circulating antibody specificities presented higher STAT4 mRNA levels compared to individuals positive for just one β cell-specific autoantibody (fold change, 1.75; p< 0.001) (Fig. 2). Thyroid-specific antibodies did not reveal an association with STAT4 expression level. Lastly, carriers of the rs7574865 minor T allele presented a tendency for increased STAT4 expression in their PBMCs compared to the wild-type homozygotes; however, the difference did not reach statistical significance (p = 0.067).
Discussion
The current study confirms an association between STAT4 polymorphism rs7574865 and the autoimmune diabetes in Polish children. According to the previous meta-analyses, this SNP is probably associated with T1D, but there seem to be considerable population differences [31, 32]. Our study is the first analysis in Slavic European populations, and one of just a few STAT4 investigations conducted in European cohorts [20, 27, 28, 38]. Studies in Cretan Greeks, British, German, and Spanish patients corroborated the association of rs7574865 with T1D [20, 27, 28, 38]. On the contrary, it was not confirmed in Brazilian and Tunisian subjects with T1D, although these analyses included relatively small cohorts and were possibly underpowered to detect the minor effect [32, 33]. Nevertheless, STAT4 consistently displays significant association with autoimmune diabetes among Asian patients [29-31]. However, only early-onset diabetes, diagnosed in children less than 7.6-years-old, was associated with rs7574865 in Korean population [29]. Similar observation of a younger age at disease onset and its more severe clinical presentation in STAT4 variant carriers was described in SLE [39]. In line, our data support that the presence of T allele at rs7574865 may be associated with a more aggressive autoimmune phenotype, characterized by an earlier T1D onset. Moreover, a dosage effect of the risk allele was noticeable with regard to the age of T1D onset in the Polish cohort.
An association of the STAT4 variant with T1D co-existing with other autoimmune conditions provides further evidence for particular susceptibility of the rs7574865 T allele carriers for the immune dysfunction. This observation is supported by the association of rs7574865 with the presence of serum aTPO and aTg antibodies in T1D subjects. Corresponding results were previously reported in a retrospective analysis of 52 T1D-associated loci, comprising STAT4, in over 8,000 T1D cases from the international case-control collection, although only aTPO thyroid antibodies were addressed in that study [38]. Former studies have corroborated the association of STAT4 polymorphism with two other major components of the autoimmune polyendocrine syndrome type 2, such as Addison’s disease and autoimmune thyroid disease [24-26, 29]. Observations in small cohorts with polyglandular autoimmunity equally confirm its implication in multiple organ-specific disorders [32, 40]. Taken together, similar to previously identified immune regulatory loci such as PTPN22 and CTLA4, STAT4 seems to be universally engaged in maintenance of the immune homeostasis [41]. In addition, according to the novel experimental data, STAT4 appears a critical factor for type 1 interferon-induced amplification of the antigen-specific cytotoxic T lymphocyte effector function [42].
PBMCs from patients with T1D presented elevated STAT4 mRNA level compared to their healthy peers. Furthermore, patients with two or three circulating autoantibodies, i.e. presumably those with more fierce autoimmune reaction, exhibited higher STAT4 expression compared to individuals positive for just one diabetes-specific autoantibody [43]. Former studies in autoimmune disease, which focused mainly on SLE, revealed that PBMCs from the rs7574865 minor allele homozygotes displayed up-regulated expression of STAT4 and IFN-α-induced genes MX-1, PKR, and IFIT-1 [15, 44]. Elevated STAT4 expression at the mRNA and protein level was also confirmed in PBMCs from the T allele carriers with early arthritis, and this observation remained significant after adjustment for age, disease activity, and glucocorticoid dose [17]. However, current analysis failed to confirm statistically significant relationship between rs7574865 and gene expression level in diabetic PBMCs. Likewise, an investigation of thymic lymphocytes from subjects with myasthenia gravis and healthy controls did not reveal association between STAT4 polymorphism and the gene expression [45]. As a matter of fact, this SNP is located in the third intron of the STAT4 gene, hence it is rather unlikely to be directly implicated in gene regulation. Rs7574865 may be just a tagger for some other SNP(s) in linkage disequilibrium, which would be the genuine causal variant(s). On the other hand, lack of significant association between this polymorphism and STAT4 expression in our study could be due to relatively small size of the currently studied cohort. The study comprised just 27 T1D cases, and might be underpowered to detect the difference between wild-type homozygotes and minor allele carriers. This is the major limitation of our analysis, which precludes drawing meaningful conclusions. Lastly, ex vivo data indicate that although rs7574865 may affect the responsiveness of pre-activated T cells, it is not associated with STAT4 protein level and phosphorylation in resting PBMCs stimulated with IL-12 or IFN-α [46]. However, following phytohaemagglutinin/IL-2 activation, CD8+ T cells from STAT4 risk allele carriers presented increased levels and phosphorylation of STAT4 protein in response to IL-12 and IFN-α, and an augmented IL-12-induced IFN-γ production in both CD8+ and CD4+ T cells [46]. Therefore, our analysis, which was limited to the resting PBMCs from the newly diagnosed T1D subjects, could not detect the relationship between STAT4 expression and genotype.
Conclusions
To summarize, this study confirms an association of the STAT4 rs7574865 variant with T1D in Polish patients. Moreover, it provides an evidence that this SNP may be connected with an earlier disease onset and increased frequency of the thyroid autoimmunity. Finally, STAT4 expression appears elevated in PBMCs from the T1D individuals, especially those with several circulating diabetes-specific autoantibodies. In contrast to most former analyses, the relationship between rs7574865 and up-regulated expression of STAT4 was not convincingly confirmed in our study, probably due to its limited power. Gene expression is of special interest as STAT4 may be implicated in mechanisms shared by several autoimmune disorders. Furthermore, its phosphorylation and activation can be effectively modulated in vitro by JAK2 and TYK2 kinase inhibitors, which provides the basis for plausible targeted therapeutic approaches in STAT4 risk allele carriers [46].