Introduction
The control of Staphylococcus aureus infection is limited because of poorly effective therapeutic strategies (Boucher et al., 2009). S. aureus is a gram-positive bacterium frequently found in human nasal mucosa either transitively or permanently (Kluytmans et al., 1997). It causes various infectious diseases ranging from mild conditions such as soft tissue infections to severe infections such as endocarditis (Diefenbeck et al., 2011). The incidence of hospital-acquired and community-acquired S. aureus infections has been rising with the increasing emergence of drug-resistant strains called methicillin-resistant S. aureus (MRSA), inducible clindamycin-resistant S. aureus (iMLSB), and constitutive clindamycin-resistant S. aureus (cMLSB) (Deresinski, 2005; Gunduz et al., 2012). Therefore, novel therapeutic compounds that are equivalent to synthetic antibiotics are being introduced urgently in the treatment of multidrugresistant (MDR) strains of S. aureus. Several investigations have reported that phytoactive compounds from herbal sources are the best options as therapies for MDR bacterial infections (Garo et al., 2007; Coutinho et al., 2008).
Syzygium cumini (L.) belonging to the family Myrtaceae (also known as java plum, black plum, jambul, and Indian blackberry) is a native species to Nepal. It is a wellknown Ayurvedic medicine. It is mainly used in the treatment of various diseases because of its antimicrobial, antidiabetic, anti-inflammatory, antitumor, antioxidant, antidiarrheal, antiviral, cardioprotective, central nervous system (CNS) stimulating, antinociceptive, antifertility, antiallergic, and antipyretic activities (Bijauliya et al., 2017). The biologically active compounds of this plant have been identified through liquid chromatography-mass spectrometry (LCMS) (Satpute et al., 2018). Hence, researchers are increasingly focusing on herbal products such as S. cumini for searching new leads to develop novel drugs against MDR microbial strains (Braga et al., 2005).
Molecular docking is a preliminary approach to screen novel therapeutic agents, and this technique is an emerging field as it reduces many complexities of the drug discovery process. Screening of lead molecules with good pharmacological properties and drug-likeness is a tedious task in the drug development process. The in silico method is an easy platform to investigate biologically active compounds with favorable ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) and drug-likeness properties (Khanna and Ranganathan, 2009). The analysis of receptor-ligand interaction is a fundamental concept of rational drug design, and prediction of such interactions by molecular docking has an increasing importance in the field of structurebased drug discovery (Lyskov and Gray, 2008).
The present study therefore evaluated the in vitro antibacterial activity of crude solvent extracts of leaves of S. cumini against the resistant strains of S. aureus. The phytocompounds present in the extracts were identified using the LCMS method, and finally, in silico analysis of the identified compounds was performed to support the results of the in vitro antibacterial activity studies.
Materials and methods
Collection of plant material
The leaves of S. cumini were collected in October 2017 from various regions of province-2, Parsa, Birgunj (27°02′31.2106″ North latitude and 084°52′27.1720″ East longitude), Nepal. The leaves of the plant were identified by a Bachelor of Ayurvedic Medicine and Surgery doctor at the Nepal Ayurveda Medical College and Teaching Hospital, Birgunj, Nepal. The leaves were thoroughly washed with water, cleaned, and dried first in shade and then in a hot air oven at 50°C until complete removal of moisture. The dried leaves were then ground in a grinder, and the coarsely powdered material was stored in an airtight container at room temperature until further use.
Plant extract preparation
The dried leaves powder (50 g) was mixed with 300 ml of respective solvents (H2O, 80% ethanol, methanol, acetone, or hexane) for 7 days with intermittent shaking at room temperature. The mixture solution was first filtered through a muslin cloth and followed by filtration through an indicator filter paper by using a Buchner funnel. The solvent was evaporated in a hot air oven at 40°C; the resulting dried crude extract was weighed and stored in a refrigerator at 4°C for further studies.
Analysis of antibacterial activity of the crude extract
Bacterial strains
To characterize the antibacterial activity of the crude extracts of S. cumini leaves prepared using five different solvents, two identified phenotypes of MLSB strains of S. aureus (one iMLSB and one cMLSB) sharing common MRSA strains were tested. The bacterial strains were isolated and identified in the Department of Microbiology, National Medical College and Teaching Hospital, Birgunj, Nepal, from seven types of 2000 clinical specimens, namely, blood, pus, body fluids, urine, swab, sputum, and urine. Staphylococcus aureus ATCC 25923 was used as a control strain. A total of 242 MRSA strains, 106 iMLSB strains, and 65 cMLSB strains of S. aureus from 310 S. aureus strains were maintained on nutrient agar slope at 4°C and activated by cultivation in nutrient broth at 37°C for 24 h before antibacterial evaluation.
Preparation of plant extract solutions
Small amounts (25, 50, 100 and 200 μg) of the five respective solvent extracts of the plant were dissolved in 1 ml of dimethyl sulfoxide (DMSO) to make a solution of four different concentrations, namely, 25, 50, 100 and 200 μg/ml). DMSO was chosen as the solvent because it has the capacity to dissolve both polar and non-polar compounds completely and it does not have any inhibitory effect on bacterial culture.
Screening of antibacterial activity of plant extracts
The antibacterial activity of the five solvent extracts was determined by the agar well diffusion technique (Nair et al., 2005). The test bacterial strains were transferred into a test tube containing 5ml of normal saline. The inoculum was prepared until the turbidity was adjusted to 0.5 McFarland standards. By using a sterile cotton swab, the prepared inoculums were inoculated onto the surface of Muller Hinton agar plates, and the uniformly swabbed plates were then allowed to dry. Five wells of 8 mm diameter were made on agar plates by using a sterile cork borer. Subsequently, 100 μl of the desired solution (25, 50, 100 and 200 μg/ml) of the extract was added to four wells. The same procedure was performed for negative control (DMSO) in the fifth well. Linezolid (30 μg/disk) was added to the plate as a positive control. The processed plates were incubated at 37°C for 24 h. The antibacterial activity was determined by measuring the inhibition zone diameter (mm) around each test bacterial strain. The experiment was conducted in triplicates.
Screening of phytocompounds present in plant extracts by LC-MS analysis
The leaf extract of S. cumini was analyzed quantitatively by LCMS at Sophisticated Analytical Instrument Facility (SAIF), Central Drug Research Institute (CDRI), Lucknow, India. The LC-MS system comprised a Waters Alliance 2695 HPLC pump, an autosampler, a vacuum degasser, and a column compartment attached with a XEVO-TQD detector with electrospray ionization (ESI) interface. The following gradient of solvents were used: water (solvent A), acetonitrile (solvent B), methanol (solvent C), and 0.1% formic acid (solvent D) were used as the mobile phase. The following gradient procedure was used: 95% B, 1.5% C, 1% D (0–1 min); 95–70% B, 1.5% C, 6% D (1–6 min); 70–40% B, 1.5% C, 6% D (6–12 min); 40% B, 1.5% C, 6% D (12–16 min); 40–20% B, 1.5% B, 6% D (16–20 min); 20% B, 1.5% C, 6% D (20–24 min); the column was re-equilibrated with 20–95% B, 1.5% C, 6% D (24–26 min) and held at 95% B, 1.5% C, and 6% D between 26 and 30 min. The injection volume was 25 μl. The XEVO-TQD#QCA1232 analysis was performed according to the following parameters of the ion sources: dual spray jet stream ESI, positive (ES+) and negative (ES-) ion mode, source temperature: 150°C, desolvation temperature: 350°C, desolvation gas flow rate: 950 (l/h), MS1 collision energy: 3, MS2 collision energy: 20, m/z range 20 to 1974 Da. The compounds were identified based on the MassLynx database and compared with literature data/survey.
Assessing drug-likeliness properties of the selected compounds
The compounds selected according to the LCMS analysis were first tested for their drug-likeliness properties based on Lipinski’s-rule of five in molecular docking analysis. The application of the rule of five or Lipinski’s rule on compounds was carried out by Molinspiration (http://www.molinspiration.com/cgi-bin/properties) which provides information about the molecular weight, logP, and the number of hydrogen bond donors and acceptors that violate the rule. The compounds not following Lipinski’s rule were eliminated from further studies (Lipinski et al., 2012).
Docking analysis
To better understand the mode of interaction and the inhibitory mechanism of the phytochemical compounds of the crude extract, docking analysis was performed using the i GEMDOCK V 2.1 package. The two drug target pathways, namely penicillin-binding protein 2a (Utsui and Yokota, 1985) of MRSA and erythromycin ribosomal methylase (ERM) (Leclercq and Courvalin, 1991) of MLSB S. aureus were used to predict the inhibition of cell wall and protein synthesis by plant-derived compounds. These two target studied proteins were retrieved from protein data bank (PDB ID: 1mwt and PDB ID: 3j7z) (Huang et al., 2015). GEMDOCK uses an empirical scoring function and an evolutionary approach. The GEMDOCK energy function consists of electrostatic, steric, and hydrogen-bonding potentials. The latter two terms use a linear model that is simple and rapidly recognizes potential complexes. The main concept of this evolutionary approach is to design multiple operators that cooperate using a family competition paradigm that is similar to a local search procedure. The parameters set in the operation of i GEMDOCK included the initial sample site (σ= 0.8 and ψ = 0.2), family competition length (L = 2), population size (N = 1000), and recombination probability (pc = 0.3). i GEMDOCK was operated through a simple scoring function. This function comprises four terms: ligand internal energy associated with torsion angles, electrostatic energy between ligand and protein, non-bonded interaction energy between ligand, and protein and penalizing solution (Epenal):
Bij– non-bonded parameter for hydrogen bonding and steric energy; rij = 4r – distance-dependent dielectric constant; rij – distance between the atoms i and j ; qi and qj – charges of atoms in ligand and receptor; lig – number of atoms in ligand; rec – number of atoms in receptor.
Analysis of ADMET properties of phytocompounds
All the identified phytocompounds were subjected to the analysis of absorption, digestion, metabolism, excretion, and toxicity (ADMET) properties by using the admetSAR server to determine the best drug candidate (Cheng et al., 2012).
Results and discussion
Extraction yield
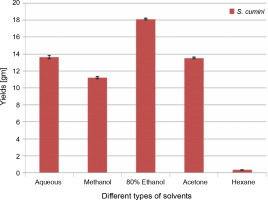
The extractive yields of crude extract of leaves of S. cumini obtained in five different solvents used were calculated, and the results are shown in Figure 1. The weight of crude extract in dry solid form ranged from 0.3 ± 0.05 to 18.1 ± 0.1 gm (w/w). The highest yield of crude extract (18.1 ± 0.1 gm) was obtained in ethanol, followed by H2O (13.6 ± 0.2 gm), acetone (13.5 ± 0.1 gm), methanol (11.2 ± 0.1 gm), and hexane (0.3 ± 0.05 gm). Statistically, the relationship between the weight of the yield of crude extract in five different solvents was significant (P < 0.05). According to our result, the yields of crude extracts varied in distilled water and organic solvents (methanol, ethanol, acetone, and hexane). The variation in yield is strongly affected by the extraction method, temperature, extraction time, solvent used, and composition of phytochemicals (McDonald et al., 2001; Turkmen et al., 2006; Ngo et al., 2017). Under the same conditions of extraction, solvent is recognized as one of the most important factors that causes variation in extraction yield. A higher extraction yield was observed in ethanolic extract, distilled water extract, and methanolic extract than in hexane and acetone extracts, indicating that the extractive yield efficacy favors highly polar solvents. These results are in line with the effect of various solvent systems used for extraction from Lathyrus maritimus L. (Chavan and Amarowicz, 2013), Helicteres hirsute (Pham et al., 2015), and some other medicinal plants (Kuppusamy et al., 2015). Significant differences were observed between extraction yield obtained in ethanol and that obtained in other solvents. This difference may be due to the higher solubility of extractable phytocomponents in ethanol than in other solvents. The variation in the solubility of phytocompounds in solvents of different polarities is also influenced by the structural differences of compounds (Felhi et al., 2016a). In a previous study, five solvents were used with different polarities and arranged in terms of dielectric constant starting from low to high values as follows: hexane (1.88) < acetone (20.7) < ethanol (25.3) < methanol (33) < water (78.4) (Maryott and Smith 1951).
Clinical specimen analysis and in vitro antibacterial assay
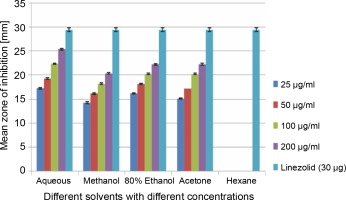
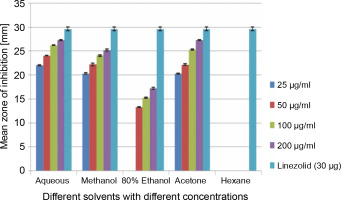
A total of 2000 clinical specimens were received, among which 310 samples showed growth of S. aureus and were used for the study purpose. The samples were categorized as blood [155 (50%)], pus [146 (47.09%)], body fluids [5 (1.61%)], urine [2 (0.65%)], swab [1 (0.32%)], and sputum [1(0.32%)]. Of the 310 S. aureus isolates, 242 (78.06%) were detected to be MRSA, 106 (34.19%) were iMLSB, and 65 (20.96%) were cMLSB. The results of the analysis of antibacterial activity of plant extracts against iMLSB and cMLSB sharing common MRSA are expressed by the size of the zones of inhibition and given in millimeters, and the results are presented in Table 1 and Figures 2 and 3. Among the two tested phenotypes of S. aureus strains (one iMLSB and one cMLSB sharing common MRSA), the different concentrations of crude plant extracts showing different antibacterial activities were qualitatively assessed by the presence or absence of inhibition zones. The aqueous, methanol, 80% ethanol, and acetone extracts of the investigated S.cumini leaves showed antimicrobial activity against both the tested bacterial strains. As shown in Table 1, the diluted aqueous extract (100 and 200 μg/ml) showed the highest antibacterial activity against both iMLSB and cMLSB strains sharing common MRSA with the inhibition zones of 22.4 ± 0.1 mm and 25.36 ± 0.15 mm, and 26.30 ± 0.10 mm and 27.30 ± 0.10 mm, respectively, whereas the 25 and 50 μg/ml extracts showed a low inhibitory effect on bacterial growth, with the inhibition zones of 17.23 ± 0.20 mm and 19.26 ± 0.20 mm, and 22.10 ± 0.10 mm and 24.10±0.10 mm, respectively. Furthermore, the methanol extract (100 and 200 μg/ml) exhibited the highest antibacterial activity against the tested strains of iMLSB and cMLSB sharing common MRSA, with inhibition zones of 18.20 ± 0.20 mm and 20.36 ± 0.15 mm, and 24.13 ± 0.15 mm and 25.26 ± 0.25 mm, respectively. The 25 and 50 μg/ml extracts showed the least inhibitory effect on iMLSB and cMLSB strains sharing common MRSA, with inhibition zones of 14.23 ± 0.25 mm and 16.16 ± 0.15 mm, and 20.36 ± 0.15 mm and 22.26 ± 0.25 mm, respectively. In contrast, the two concentrations (100 and 200 μg/ml) of 80% ethanol extract showed antibacterial activity against iMLSB and cMLSB strains sharing common MRSA with the inhibition zones of 20.23 ± 0.15 mm and 22.23 ± 0.15 mm, and 15.30 ± 0.10 mm and 17.30 ± 0.20 mm, respectively. Lower concentrations (25 and 50 μg/ml) of the extract showed inhibitory effect with zones of inhibitions of 16.20 ± 0.10 mm and 18.16 ± 0.11 mm, and 0 and 13.30 ± 0.10 mm, respectively. Moreover, the acetone extract at 100 and 200 μg/ml concentrations exhibited high antimicrobial effect on iMLSB strain (20.23 ± 0.15 mm and 22.23 ± 0.20 mm) and cMLSB strain (25.36 ± 0.15 mm and 27.34 ± 0.15 mm) sharing common MRSA, whereas the extract at 25 and 50 μg/ml concentrations showed lesser inhibitory activity with inhibition zones of 15.10 ± 0.10 mm and 17.16 ± 0.05 mm for iMLSB and 20.30 ± 0.10 mm and 22.23 ± 0.15 mm for cMLSB. The aqueous extract of S. cumini leaves showed the highest antibacterial activity with a higher zone of inhibition against both the studied strains of S. aureus (iMLSB and cMLSB sharing common MRSA). All the five extracts with its zones of inhibition were compared with standard antibiotic linezolid (30 μm/disc), and the results are presented in Table 1. All the studied strains were sensitive to linezolid, with the highest zone of inhibition of 29.43 ± 0.4 mm against iMLSB and 29.66 ± 0.35 mm against cMLSB. The antibacterial effect of the plant extract was comparable to that of the positive control antibiotic, as shown in Table 1. These results indicate that the extract of S. cumini leaves is a potential antibacterial agent. The antimicrobial activity of S. cumini leaves extract is probably caused by the presence of tannins and other phenolic constituents. S. cumini is very rich in gallic and ellagic acid polyphenol derivatives (Bajpai et al., 2005; Abhishek and Vinod, 2011). In this regard, Shafi et al. (2002) reported that the leaves of S. cumini showed antimicrobial activity against different strains of bacteria, such as S. aureus, Salmonella typhimurium, Pseudomonas aeruginosa, Escherichia coli, Bacillus subtilis, and Bacillus sphaericus. Shyamala and Vasantha (2010) also mentioned that the S. cumini leaf extract showed antibacterial activity against E. coli and S. aureus. Benarjee and Narendhirakannan (2011) demonstrated the antimicrobial activity S. cumini ethanolic seed extract (250 mg/ml), with inhibition zones of 18–22 mm against gram-negative bacteria and 20–23 mm against gram-positive bacteria.
Table 1
Inhibition zone in mm of solvent extracts of Syzygium cumini leaves against bacterial strains
Profiling of phytocompounds in S.cumini
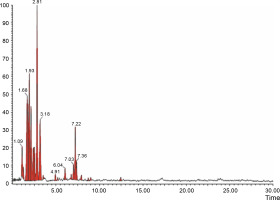
The aqueous extract that exhibited the highest antibacterial effect was analyzed and profiled by the LC-MS analysis in positive and negative ionization modes to qualitatively characterize active compound constituents. The base peak chromatogram is shown in Figure 4 (positive ionization mode) and Figure 5 (negative ionization mode). The characterization of compounds was performed based on the mass spectrum obtained in MS-ESI and compared with the previous literature reports (Zehl et al., 2011; Costa et al., 2015; Wang et al., 2015). The results of the LC-MS analysis enabled tentative identification of 20 different compounds, as shown in Table 2, with their retention time, molecular weight, monoisotropic mass, and molecular formula. The 20 identified compounds included eight phenolic compounds: caffeic acid, 3-(3-hydroxy phenyl) propionic acid, xanthoxylin, ferulic acid, quinic acid, diferulic acid, methyl gallate, and gallic acid; eight flavonoids: astragalin, cianidanol, butin, kaempferide, 4′-hydroxyflavan, taxifolin, isoquercetin, and 3,5,7,4′-tetrahydroxy-6-(3-hydroxy-3-methylbutyl) flavone; one terpenoid: cedrol; one coumarin: 6-O-feruloyl-D-glucose; and two organic acids: palmitic acid and punicic acid.
Table 2
Identification of active compounds in S. cumini leaves water extract by LCMS
| S. No | Rt [min] | MolWt | ESI mode | Monoisotropic mass | Chemical formula | Compounds | References |
|---|---|---|---|---|---|---|---|
| 1 | 6.14 | 386 | [M+H]+ | 386.100168 | C20H18O8 | diferulic acid | Ramya et al., 2012 |
| 2 | 8.58 | 184 | [M+H]+ | 184.037173 | C8H8O5 | methylgallate | Ramya et al., 2012 |
| 3 | 2.18 | 290 | [M+H]+ | 290.079038 | C10H12O4 | cianidanol | Chhikara et al., 2018 |
| 4 | 9.42 | 273 | [M+H]+ | 272.068473 | C15H12O5 | butin | Ramya et al., 2012 |
| 5 | 15.82 | 300 | [M+H]+ | 300.063388 | C16H12O6 | kaempferide | Ramya et al., 2012 |
| 6 | 3.27 | 226 | [M+H]+ | 226.09938 | C15H14O2 | 4′-hydroxyflavan | Bijauliya et al., 2017 |
| 7 | 20.07 | 304 | [M+H]+ | 304.058303 | C15H12O7 | taxifolin | Singh et al., 2018 |
| 8 | 10.63 | 257 | [M+H]+ | 256.24023 | C16H32O2 | palmitic acid | Ayyanar et al., 2012 |
| 9 | 22.28 | 278 | [M+H]+ | 278.224580204 | C18H30O2 | punicic acid | Chagas et al., 2015 |
| 10 | 14.50 | 222 | [M+H]+ | 222.198365 | C15H26O | cedrol | Satpute et al., 2018 |
| 11 | 1.67 | 180 | [M+H]- | 180.042259 | C9H8O4 | caffeic acid | Ramya et al., 2012 |
| 12 | 1.74 | 166 | [M+H]- | 166.062994 | C9H10O3 | 3(3-hydroxyphenyl)propanoic acid | Singh et al., 2018 |
| 13 | 1.92 | 196 | [M+H]- | 196.073559 | C10H12O4 | xanthoxylin | Chagas et al., 2015 |
| 14 | 2.50 | 194 | [M+H]- | 194.057909 | C10H10O4 | ferulic acid | Chhikara et al., 2018 |
| 15 | 2.82 | 192 | [M+H]- | 192.063388 | C7H12O6 | quinic acid | Singh et al., 2018 |
| 16 | 7.93 | 448 | [M+H]- | 448.100561 | C21H20O11 | astragalin | Bijauliya et al., 2017 |
| 17 | 3.52 | 356 | [M+H]- | 356.1107 | C16H20O9 | 6-O-feruloyl-D-glucose | Ramya et al., 2012 |
| 18 | 3.18 | 170 | [M+H]+/[M+H]- | 170.021523 | C7H6O5 | gallic acid | Timbola et al., 2002 |
| 19 | 7.21 | 464 | [M+H]+/[M+H]- | 464.095476 | C21H20O12 | isoquercetin | Jagetia, 2017 |
| 20 | 7.33 | 372 | [M+H]+/[M+H]- | 372.120903 | C20H20O7 | 3,5,7,4′-tetrahydroxy-6-(3-hydroxy-3-methylbutyl)flavone | Chagas et al., 2015 |
Fig. 4
LCMS chromatogram of aqueous extract of Syzygium cumini leaves in positive mode showing peaks with identified compounds listed in Table 2
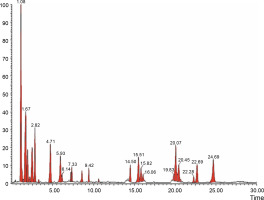
Fig. 5
LCMS chromatogram of aqueous extract of Syzygium cumini leaves in negative mode showing peaks with identified compounds listed in Table 2
The bioactivities of some compounds identified from the extract of S. cumini leaves in this study have been documented in previous studies (Ayyanar et al., 2012; Ramya et al., 2012; Chagas et al., 2015; Bijauliya et al., 2017; Jagetia, 2017; Chhikara et al., 2018; Singh et al., 2018). The bioactive phenolic compounds identified in the leaves of S. cumini showed defense responses in the plants. Phenolic metabolites also play an important role in other processes, for example, incorporation of attractive substances to accelerate pollination, coloring for camouflage, and defense against herbivores as well as antibacterial and antifungal activities (Alasalvar et al., 2001; Acamovic and Brooker, 2005; Edreva et al., 2008). The flavonoids identified in S. cumini are an important class of natural products, and they belong to a class of plant secondary metabolites with a polyphenolic structure. They are associated with a broad spectrum of health-promoting effects and are an indispensable component in various medicinal, pharmaceutical, and cometic applications (Panche et al., 2016). This is because of their antimicrobial, antioxidative, anti-inflammatory, anticarcinogenic, and antimutagenic properties coupled with their capacity to prevent and treat different diseases (Panche et al., 2016). Cedrol, one of the compounds belonging to the terpenoid group, has been reported for its pharmacological activities, and it is used as a sedative, anti-inflammatory, antibacterial, and antifungal agent (Dayawansa et al., 2003; Satpute and Vanmare, 2018). Coumarin, 6-O-feruloyl-D-glucose, has strong antioxidant, antimicrobial, and probiotic properties (Ou and Sun, 2014). The two organic acids (palmitic acid and punicic acid) play a role in maintaining homeostatic balance and are implicated in different pathophysiological conditions such as atherosclerosis, neurodegenerative diseases, and cancer (Carta et al., 2017; Aruna et al., 2016).
Drug-likeliness properties of the selected compounds
The drug-likeliness properties of the 20 identified compounds, namely caffeic acid, 3-(3-hydroxy phenyl) propionic acid, xanthoxylin, ferulic acid, quinic acid, diferulic acid, methyl gallate, gallic acid, astragalin, cianidanol, butin, kaempferide, 4′-hydroxyflavan, taxifolin, isoquercetin, 3,5,7,4′-tetrahydroxy-6-(3-hydroxy-3-methylbutyl) flavones, cedrol, 6-O-feruloyl-D-glucose, palmitic acid, and punicic acid, from S. cumini are given in Table 3. To determine the drug potential of all the ligands, Lipinski’s rule of five (RO5) (Hari, 2019) as it is considered to be the potential technique was used for in silico studies. Molinspiration is a computer-based technique in which a number of properties (such as lipophilicity in terms of log P, number of hydrogen bond acceptors, number of hydrogen bond donors, molecular weight, and number of violations) of the compounds have been characterized (Lipinski et al., 2012). Our results showed that the molecular weight of the studied compounds were in the range of 170–464. The molecular weight values were within the reference values in RO5, i.e., ≤ 500. The hydrogen bond acceptor (Ha) and hydrogen bond donor (Hd) of compounds were calculated and determined to be < 10 (Ha) and < 5 (Hd), respectively, in RO5 (Baell et al., 2013). Our results showed that all the 20 tested compounds were in the range of 1–8, which met the reference value of < 10 for hydrogen bond acceptor. For hydrogen bond donor (Hd), all the compounds were found in the reference value of < 5, except for cianidanol, gallic acid, quinic acid, 3,5,7,4′-tetrahydroxy-6-(3-hydroxy-3-methylbutyl) flavones, 6-O-feruloyl-D-glucose, isoquercetin, astragalin, methyl gallate, butin, kaempferide, and taxifolin. Log P is the parameter that is used to assess the lipophilicity of the compounds. Our results showed that compounds tested in this study met the requirements as lipophilic compounds, with log P < 5, and therefore, they were able to penetrate the cell membrane. Thus, 18 of the 20 tested compounds fulfilled RO5, while two compounds (isoquercetin and astragalin) did not fulfill RO5. Hence, these two compounds were excluded from the docking analysis.
Table 3
Lipinski properties of plant compounds analyzed using molinspiration
Molecular docking
The 18 tested compounds were docked using penicillin-binding protein 2a (PBP2a) and erythromycin ribosomal methylase (ERM) target proteins. The PBP2a enzyme plays a crucial role in β-lactam resistivity, and it is considered as the prime target for MRSA infection. PBP2a is a high-molecular-weight class B penicillin binding protein (PBP) and is found only in MRSA. Functionally, PBP2a is a unique transpeptidase that is not inhibited by β-lactam antibiotics; hence, it can continue with peptidoglycan crosslinking even in the presence of these antibiotics (Fishovitz et al., 2014). The ERM proteins methylate the single adenine residue within 23S rRNA to reduce the affinity of antibiotics to a region around the peptidyl transferase center, thereby conferring resistance to macrolide, lincosamide and streptogramin B (MLSB) antibiotics in various microorganisms, especially in iMLSB and cMLSB phenotypic strains of S. aureus (Li et al., 2017). The ERM proteins are composed of two domains: a catalytic domain and a substrate-binding domain. The larger N-terminal catalytic domain exhibits a typical α/β/α sandwich architecture that contains the S-adenosyl-L-methionine-binding site and the smaller C-terminal domain consists of three α-helices that function as an rRNA-binding domain (Stsiapanava and Selmer, 2019).
Docking studies are used at several stages of drug discovery to find ligand-receptor interactions and to rank the compounds based on the binding energies or fitness score (Emran et al., 2015). The binding scores of the 18 compounds to PBP2a and ERM proteins are shown in Table 4. For PBP2a, the docking scores ranged from –65.1 to –102.4 kcal/mole. The binding energies of the compounds diferulic acid and taxifolin did not differ significantly from that of the reference antibiotic linezolid (–103 kcal/mole). For the ERM protein, the docking scores ranged from –64.4 to (–103.2 kcal/mole). The compound taxifolin showed a higher binding energy than the reference antibiotic linezolid (–102.6 kcal/mole). The protein-ligand interactions between the target proteins, the higher ranking compounds (in terms of binding energy), and linezolid (the standard antibiotic) are presented in Figures 6 and 7. The interaction and predicted pose between diferulic acid and PBP2a (Fig. 6B) showed that the interactions were stabilized by van der Waals forces through Tyr 446, Asn 464, His 583, Gly 599, Thr 600, Gln 521, Glu 602, and Met 641 residues. The interactions between taxifolin and ERM were stabilized by hydrogen bonds formed by amino acid residues: Ser 403, Tyr 446, Ser 462, Asn 464, His 583, Ser 598, Gly 599, Thr 600, Ala 642, and Ser 643,and van der Waals interactions involving the residues Thr 444, Tyr 446, Thr 600, and Met 641 (Fig. 7B). For the standard drug linezolid, the predicted binding pose with PBP2a (PDB ID: 1mwt) showed interactions stabilized by hydrogen bonds with Ser 403, Gln 521, Gly 599, and Thr 600 and van der Waals interactions with Tyr 446, Thr 600, and Met 641 (Fig. 6A). The standard antibiotic also showed a large interaction with the ERM protein (PDB ID: 3j7z) stabilized by hydrogen bonds with Ser 403, Gln 521, Gly 599, and Thr 600 and van der Waals interaction involving Tyr 446, Thr 600, and Met 641 residues (Fig. 7A). Because natural compounds are considered to be safe, nontoxic, and less prone to side effects (Rani et al., 2014), in the present study, naturally occurring phytochemicals from different plant sources were selected and validated for PBP2a inhibition in MRSA infection. Molecular docking analysis suggested that diferulic acid had a higher specificity toward the PBP2a binding site and thus caused inhibition of MRSA by inhibiting the biosynthesis of its cell wall (Rani et al., 2014). It is notable that the compound taxifolin exhibited antibacterial activity by hindering with the function of ERM in MLSB strains of S. aureus. Taxifolin has been reported to play a role in the inhibition of microorganisms and may be important in increasing bioaccessibility and bioavailability; thus, it can be used as an effective drug (Raj et al., 2017). Taxifolin can enhance the effectiveness of traditional antibiotics such as ceftazidime and levofloxacin in vitro; thus, a combination therapy involving taxifolin and traditional antibioticscan be provided to patients with MDR S. aureus and related infections (An et al., 2011). This is a strong indication that these compounds could have contributed significantly to the observed anti-MRSA and anti-MLSB activity of the extracts of S. cumini leaves. This could be attributed to the structural similarity between the conventional substrates or ligands of the target proteins and the compounds isolated from the plants.
Table 4
iGEMDOCK docking score for compounds (ligands) from aqueous extract of Syzygium cumini leaves [Kcal/mol]
Fig. 6
Predicted docking pose of (A) linezolid (reference antibiotic) and (B) diferulic acid lie within the active site of target protein (PDB ID: 1mwt). The pink color represents the corresponding ligand molecule and the green color represents the corresponding reference. Green and gray color represents the amino acids involved in hydrogen bonding and van der Walls interaction respectively
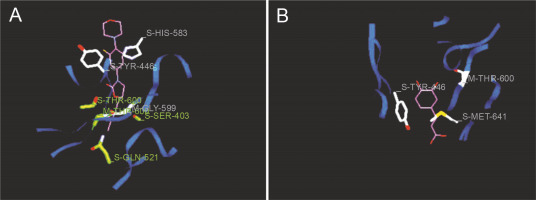
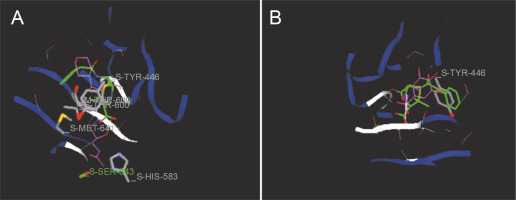
Fig. 7
Predicted docking pose of (A) linezolid (reference antibiotic) and (B) taxifolin lie within the active site of target protein (PDB ID: 3j7z). The pink color represents the corresponding ligand molecule and the green color represents the corresponding reference. Green and gray colors represent the amino acids involved in hydrogen bonding and van der Walls interaction respectively
Prediction of pharmacokinetic properties of the selected compounds
The ADMET properties of the selected compounds, namely, caffeic acid, 3-(3-hydroxy phenyl) propionic acid, xanthoxylin, ferulic acid, quinic acid, diferulic acid, methyl gallate, gallic acid, cianidanol, butin, kaempferide, 4′-hydroxyflavan, taxifolin, 3,5,7,4′-tetrahydroxy-6-(3-hydroxy-3-methylbutyl) flavones, cedrol, 6-O-feruloyl-D-glucose, palmitic acid, and punicic acid, were studied to determine their eventual fate in the body (Shin et al., 2016). For this analysis, the admetSAR server was used (Cheng et al., 2012). Human intestinal absorption (HIA), blood-brain barrier (BBB) penetration, acute oral toxicity, AMES toxicity, and carcinogenicity were determined. The results revealed that all the tested compounds were predicted to be non-AMES toxic or noncarcinogenic or nonmutagenic agents. These results imply greater chemical inactivation or prevention of damage to DNA by the chemical compounds of S. cumini, which is accordance to with the results of Edenharder et al., (1993), who examined the effect of structurally related flavonoids and related compounds and showed distinct structure-activity relationship and protective activity. Our results also revealed that the compounds had good HIA and BBB properties. Compounds for oral administration should be well absorbed in the gastrointestinal tract (GIT) for optimal pharmacokinetics. The BBB is a very important barrier that limits the penetration of compounds into the CNS (Sweeney et al., 2019). The acute oral toxicity of compounds is analyzed by LD50, which is defined as the amount of material required to cause mortality of 50% of a group of test animals (Ammar, 2017). For both economical and ethical reasons, non-animal-based prediction of LD50 was performed by the ADMET analysis. The acceptable range of LD50 for an ideal drug candidate corresponding to its acute oral toxicity is ≥ 50 mg/kg, and the other categories are ≤ 500 mg/kg (category II), ≥500mg/kg but ≤ 5000 mg/kg (category III), and ≥ 5000 mg/kg (category IV). Two tested compounds (butin and taxifolin) were in category II, 11 compounds (diferulic acid, xanthoxylin, quinic acid, gallic acid, 4′-hydroxyflavan, 3,5,7,4′-tetrahydroxy-6-(3-hydroxy-3-methylbutyl)flavone, 6-O-feruloyl-D-glucose, methyl gallate, cedrol, kaempferide, and 3-(3-hydroxyphenyl) propanoic acid) were in category III, and 5 compounds (caffeic acid, cianidanol, ferulic acid, palmitic acid, and punicic acid) were in category IV. Assessment of acute toxicity of compounds is required to determine their adverse effects that might occur due to accidental or deliberate short-term exposure and also serve as a guide in dose selection for long-term toxicity studies as well as for other studies involving the use of animals (Maheshwari and Sheikh, 2016; Erhirhie et al., 2018).
Conclusions
Our study demonstrated that the solvent extract of S. cumini leaves possessed significant and dose-dependent anti-MRSA and anti-MLSB activities, which supports the traditional use of this plant in folk medicine. The positive result regarding the antibacterial activities increases the value of this plant. Collectively, the results of the present study support the ethnomedicinal use of S. cumini for the management of various infectious diseases. Furthermore, the different potential bioactive compounds identified by the LC-MS analysis in the S. cumini leaf extract showed promising binding affinity toward two proteins (PBP2a and ERM) in molecular docking analysis, and their drug-likeness characteristics were demonstrated through the ADMET experiment. The docking analysis result showed that the compounds diferulic acid and taxifolin yielded better results in terms of binding energy toward MRSA and MLSB resistant S. aureus strains, respectively. With the continuous increase in antibiotic resistance, further studies of these compounds and its derivatives could be a promising and effectual antibiotic lead against the emerging MRSA and MLSB resistant strains of S. aureus.