Introduction
The activation of antigen-specific T cells is key for developing adaptive immunity [1]. The T-cell receptor (TCR)-CD3 complex, which is expressed on T-cell membranes, is capable of integrating and transducing signals engaged by peptide-major histocompatibility complex (MHC) complexes expressed on antigen-presenting cells (APCs) [2]. Each TCR-CD3 complex contains one CD3γ and CD3δ chain and two CD3ε and CD3ζ chains [3].
Recent evidence has demonstrated that CD3ζ plays a vital role in multiple autoimmune, inflammatory, and malignant diseases [4-6]. The CD3ζ gene is regulated at the transcriptional, posttranscriptional, and posttranslational levels [7].
We previously demonstrated that different CD3ζ 3’ untranslated region (3’-UTR) alternatively spliced isoforms regulate the CD3ζ mRNA expression level in T cells in patients with aplastic anaemia (AA) and chronic myeloid leukaemia (CML) [8, 9]. These results revealed that a posttranslational mechanism was involved in regulating CD3ζ expression.
MicroRNAs (miRNAs) are endogenous, small non-coding RNA molecules that act as posttranscriptional regulators. miRNAs specifically pair with complementary sites in 3’-UTRs of target mRNAs to block translation or increase mRNA degradation [10]. Recently, strong evidence has suggested the important role of miRNAs in T-cell immunity [11]. miR-202-3p plays a role in posttranscriptional regulation of CD3ζ in pancreas-infiltrating T cells of non-obese diabetic mice [12]. It is well known that each gene is target regulated by many miRNA; thus, we explored whether there are other miRNAs involved in the regulation of CD3ζ.
In this study, miRNA target gene databases were used to predict potential miRNAs that target CD3ζ mRNA. The predicted miRNA miR-214, which regulates CD3ζ gene expression, was verified in the MOLT-4 cell line, demonstrating that miR-214 may be a key regulator of CD3ζ.
Material and methods
Cell cultures
Acute lymphoblastic leukaemia MOLT-4 cells were maintained in RPMI-1640 medium (Gibco, USA) supplemented with 10% foetal bovine serum (Gibco, USA) and antibiotics (100 U/ml penicillin and 100 U/ml streptomycin).
Human embryonic kidney (HEK 293T) cells were maintained in GlutaMAX High Glucose DMEM medium (Gibco, USA) supplemented with 10% foetal bovine serum (Gibco, USA) and antibiotics (100 U/ml penicillin and 100 U/ml streptomycin).
All cell lines were incubated at 37°C in a humidified atmosphere of 5% CO2.
Construction of CD3ζ 3’-UTR luciferase reporter vectors
The wild type CD3ζ 3’-UTR, which includes a conserved putative miR-214 binding site (1050-1056 bp), was amplified by polymerase chain reaction (PCR). The PCR products were cloned into the pGEM-T Easy Vector (Promega, USA).
After cloning, a Fast Site-Directed Mutagenesis Kit (TIANGEN, China) was used to generate a mutation in the CD3ζ 3’-UTR, according to the manufacturer’s protocol. The site-directed mutagenesis primers were as follows: forward: 5’-TCGAGTGTGTCTGAGTGGCTTCACTCACGACGTAAATTTGGCTTCTGTTG TCGTTT-3’ and reverse: 5’-AAACGACAACAGAAGCCAAATTTACGTCGTG AGTGAAGCCACTCAGACACAC-3’ (mutations are underlined). The wild type and mutant CD3ζ 3’-UTR were subcloned from the pGEM-T Easy Vector into the psiCHECK-2 plasmid (Promega, USA) using the following primers: forward: 5’-CCGCTCGAGCAGCCAGGGGATTTCACCACTCAAAG-3’ (Xho I), and reverse: 5’-AGCTTTGTTTAAACCCCTAGTACATTGACGGGTTTT TCC TG-3’ (Pme I). Successful construction of the psiCHECK-2 plasmids including the wildtype, and mutant CD3ζ 3’-UTRs (CHK2-CD3Z-WT and CHK2-CD3Z-MU) was verified by sequencing.
Luciferase assays
HEK293T cells were cultured in 12-well plates (2.5 × 105/well) to approximately 80% confluence after 24 hours. The cells were then cotransfected with the CHK2-CD3Z-WT or CHK2-CD3Z-MU plasmids plus a miR-214 mimic or negative control (NC) for 48 hours. The miR-214 mimics or negative control were synthesised by Guangzhou RiboBio (RiboBio, China). The Dual-Glo Luciferase Assay System (Promega, USA) was used to evaluate the relative activity of Firefly and Renilla luciferase according to the manufacturer’s protocol. Transfections were performed in duplicate and repeated three times.
Transfection and Western blotting
The miR-214 mimic or negative control (RiboBio, China) were transfected into MOLT-4 cells using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s protocol. Transfections were performed in duplicate and repeated three times.
The miR-214 mimic or negative control were transfected into MOLT-4 cells for 72 hours, and the protein levels were analysed by Western blotting. Briefly, total cellular protein was extracted with RIPA lysis buffer (Cell Signalling Technology, USA), and protein concentrations were determined using the BCA assay. Protein aliquots (100 μg) were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), and the separated proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane. The PVDF membranes were blocked and incubated with antibodies directed against CD3ζ (1 : 500) or β-actin (1 : 1,000) followed by mouse anti-human secondary antibodies. All antibody reagents were from Cell Signal Technology with the exception of the CD3ζ antibody, which was from Abcam.
Real-time relative quantitative PCR (qRT-PCR) for CD3ζ gene expression
A miR-214 mimic or negative control was transfected into MOLT-4 cells. After 48 hours, RNA extraction and cDNA synthesis were performed according to the manufacturer’s instructions. qRT-PCR using the SYBR Green I method was then used to examine the CD3ζ gene expression level with β2M serving as an internal control. The CD3ζ and β2M primer sequences and PCR conditions were described previously [13]. The 2(–ΔCT) method was used to analyse the CD3ζ gene expression level relative to control.
Results
Predicting a potential miRNA that targets CD3ζ using bioinformatics databases
In this study, we predicted a potential miRNA that targets CD3ζ mRNA using bioinformatics databases. First, miR-214 was identified by three bioinformatics tools: TargetScan (http://www.targetscan.org), PicTar (http://www.pictar.mdc-berlin.de), and microRNA (http://www.microRNA.org). By coincidence, miR-214 was also predicted and concluded to target CD3ζ using a bioinformatics analysis tool and reviewing reports [14]. Therefore, miR-214 was chosen for experimental validation.
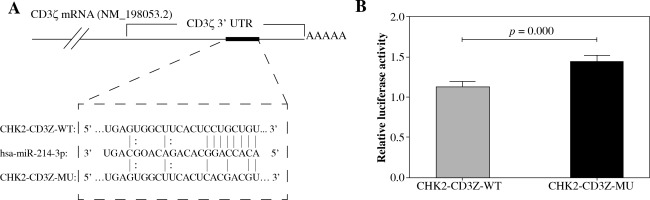
TargetScan predicted a binding site for miR-214 in the CD3ζ 3’-UTR (1050-1056 bp, NM_198,053.2). To determine whether miR-214 could specifically pair with its putative complementary site in the CD3ζ 3’-UTR, a psiCHECK-2 luciferase reporter expressing the wild type CD3ζ 3’-UTR (CHK2-CD3Z-WT) or a mutant CD3ζ 3’-UTR (CHK2-CD3Z-MU) containing point mutations in the putative miR-214 binding site was constructed and transfected into HEK293T cells together with miR-214 mimics. The relative luciferase activity of the CHK2-CD3Z-WT-transfected cells was significantly decreased (by 21.82%) compared with the CHK2-CD3Z-MU-transfected cells (Fig. 1). The results demonstrated that miR-214 specifically binds to the binding site predicted in the CD3ζ 3’-UTR, resulting in lower relative luciferase activity.
Fig. 1
Validation of miR-214 targeting the CD3ζ 3’-UTR by luciferase assay. A) The predicted binding site for miR-214 in the CD3ζ 3’-UTR. The red letters indicate mutation sites in the CD3ζ 3’-UTR. B) The miR-214 mimic (100 nM) or negative control were transfected with CHK2-CD3Z-WT or CHK2-CD3Z-MU reporter plasmids into HEK293T cells. After 48-h co-transfection, the luciferase activity was measured with the Dual-Glo Luciferase Assay System. The relative luciferase activity was significantly decreased in the CHK2-CD3Z-WT group
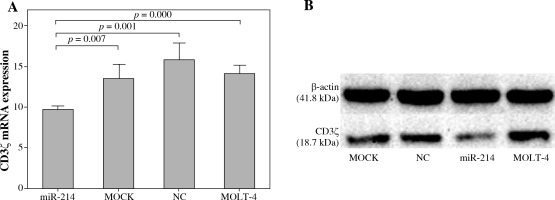
miR-214 regulated CD3ζ expression in T-cell line
We further investigated whether miR-214 could regulate CD3ζ expression in T cells by transfecting a miR-214 mimic into MOLT-4 cells. The effects of miR-214 on CD3ζ mRNA and protein expression were assessed using qRT-PCR and Western blotting. As shown in Figure 2, the CD3ζ mRNA expression level in the miR-214-mimic transfected cell group (9.76 ±0.45) was significantly lower than that in the MOLT-4, MOCK, and negative control groups (14.24 ±1.00, 13.56 ±1.81, and 15.98 ±2.04, respectively). Accordingly, Western blotting revealed that miR-214 reduces the CD3ζ protein level. These results were in accordance with the dual luciferase reporter assay findings and confirmed that miR-214 regulates CD3ζ.
Discussion
T cells play a central role in cell-mediated immune responses. T cells respond to antigens via the TCRs that recognise specific antigen peptides on APC cells. The TCR-associated CD3 complex triggers intracellular signalling cascades. The CD3 complex possesses 10 immune receptor tyrosine-based activation motifs (ITAMs), which are essential for TCR signal transduction and T-cell activation [15, 16]. Each CD3ζ chain contains three ITAMs. The structural characteristics of CD3ζ suggest that CD3ζ plays a unique role in the TCR signalling pathway. Up- and down-regulation of CD3ζ was found in different diseases with abnormal T-cell immune statuses, including autoimmune diseases such as aplastic anaemia, which overexpress CD3ζ in T cells, and cancer patients with T-cell immunosuppression, who have significantly decreased CD3ζ expression [8, 9, 17]. Reasons for the abnormal CD3ζ expression may be the transcription factor E-74-like factor (Elf-1), ubiquitination, granzyme-B, and caspase-mediated degradation [18-21]. Several studies have indicated that the CD3ζ 3’-UTR has regulatory elements that affect CD3ζ expression in T cells in patients with systemic lupus erythematosus (SLE) [22, 23]. Our previous studies have found that different CD3ζ 3’-UTR alternative splicing isoforms might regulate the CD3ζ mRNA expression level in T cells in patients with AA and CML [8, 9]. However, the mechanism involved in regulating the CD3ζ 3’-UTR remains unclear. It has been shown that miRNAs play an important role in T-cell immune homeostasis by directly targeting the 3'-UTRs of mRNAs that mediate gene expression [24]. A recent study has shown that miRNAs can modulate TCR signalling molecules [25]. Meanwhile, miR-202-3p was identified regulating CD3ζ in pancreas-infiltrating T cells of non-obese diabetic mice [12]. Taking into account the important function of miRNAs in T-cell immunity, in this work, we sought to ascertain whether other miRNAs were involved in regulation of CD3ζ.
In this study, luciferase activity assays revealed that miR-214 directly binds the CD3ζ 3’-UTR to downregulate luciferase activity. miR-214 mimic down-regulated CD3ζ expression in mRNA and protein level in T-cell line. These results revealed that miR-214 regulates CD3ζ.
Targets of miRNA-214 are complex and mediate diverse processes such as differentiation, senescence, angiogenesis, cell migration, and virus replication; it also acts as a tumour suppressor or oncogene in different cancers [26, 27]. In this study, we found that miR-214 might have an additional function involving the regulation of T-cell activation. However, it has also been reported that there is increased miR-214 expression in activated T cells. In this case, it was thought that the role of miR-214 might be to promote T-cell activation in C57BL/6 mouse T cells by targeting PTEN [28]. Therefore, it is thought that the contrasting effects of miR-214 on T-cell activation might be related to different cell types and disease statuses. This hypothesis is similar to findings with miR-155, which was demonstrated to possess multiple opposing functions in various cell types, including promoting regulatory T cells and T-effector function [29]. However, further investigation is needed to determine the function of miR-214 in different T-cell subsets with different T-cell immune statuses and diseases, and whether miR-214 regulates different genes in T cells through its complex target networks.
Conclusions
In conclusion, we identified for the first time that miR-214 targets CD3ζ expression in MOLT-4 cells, implying that miR-214 might negatively regulate T-cell activation by targeting CD3ζ. This finding might provide a novel approach for targeted regulation of the CD3ζ expression level to control the T-cell activation status. Further investigation will focus on the regulatory mechanisms of miR-214 in haematological diseases with different T-cell immune statuses.