Introduction
Neutrophil extracellular traps (NETs) consist of chromatin filaments coated with histones, proteases, and other granular and cytosolic proteins [1]. Recognized as an important neutrophil feature, NET release contributes to microbial capture, immobilization, and killing, but overzealous NET release can significantly damage host tissue [2-4]. Previous studies have demonstrated the substantial contribution of NETs to mortality across multiple disorders, including infectious diseases, autoimmune diseases, cancer, and thrombosis [4, 5]. However, the mechanisms underlying deleterious NET effects are not yet fully understood.
Although in vitro incubation of cells with NETs is commonly used to evaluate the impact of NETs on cellular function, it is unclear which human NET isolation and handling protocol is superior for this application [6-11]. Therefore, we assessed the efficacy and efficiency of different available protocols.
Material and methods
Ethical approval and participant consent
This study was approved by the Ethics Committee of Wuxi Children’s Hospital (WXHC2020-03-003). Six children were enrolled as healthy volunteers, and informed consent was provided by the parents of the six children.
Protocol selection and modification
Four previously described NET isolation and handling protocols were selected and labeled A-D (Table 1) [7-9, 12, 13]. The original protocols specify distinct neutrophil culture conditions and NETosis-activating stimuli, but since the specific goal of the present study was to evaluate NET isolation performance, all other differences were minimized: a single neutrophil isolation method was used, culture conditions were identical, and only phorbol 12-myristate 13-acetate (PMA) (FMS, China, catalog # FMS-FZ207) was used to stimulate NETosis.
Table 1
Protocols for NET isolation and handling
| Protocol | NET isolation procedure | Reference(s) |
|---|---|---|
| A | 1. Gently aspirate and discard culture supernatant. 2. Using 15 ml of cold PBS (lacking Ca2+ and Mg2+) wash each sample – wash the bottom of each dish by pipetting PBS on the bottom of the dish to detach all adherent material – into a 15 ml conical tube. 3. Centrifuge for 10 min at 450 × γ and 4oC to pellet neutrophils, leaving NETs suspended in the supernatant. 4. Divide the supernatant in each tube into 1.5 ml micro-centrifuge tubes and centrifuge for 10 min at 18,000 × γ and 4oC to pellet NETs. 5. Discard supernatants, recombining all pellets deriving from a common 15 ml conical tube with 5 ice cold RPMI. | [12] |
| B | 1. Gently aspirate and discard culture supernatant. 2. Using 5 ml of fresh RPMI, wash each sample – wash the bottom of each dish by pipetting medium on the bottom of the dish to detach all adherent material – into a 15 ml conical tube. 3. Centrifuge for 5 min at 1500 rpm/450 × γ to pellet neutrophils, leaving NETs suspended in the supernatant. | [7] |
| C | 1. Gently aspirate and discard culture supernatant. 2. Add 5 ml of fresh RPMI containing 10 U/ml restriction enzyme AluI and incubate neutrophils for 30 min at 37oC. 3. Wash the bottom of each dish by pipetting medium on the bottom of the dish and collect each supernatant into a 15 ml conical tube. 4. Centrifuge for 5 min at 300 × γ to pellet any remaining neutrophils, leaving NET fragments suspended in the supernatant. | [8, 13] |
| D | 1. Wash the bottom of each dish by pipetting medium on the bottom of the dish, collect all neutrophils and supernatants into a 15 ml conical tube. 2. Centrifuge for 5 min at 500 × γ to pellet neutrophils. 3. Transfer supernatants to fresh 1.5 ml micro-centrifuge tubes. 4. Centrifuge for 10 min at 18,000 × γ and 4oC to pellet NETs. 5. Discard supernatants, recombining all pellets deriving from a common 15 ml conical tube with 5 fresh RPMI. | [9] |
Peripheral blood neutrophil isolation
Neutrophils were isolated from freshly collected blood samples using a Neutrophil Isolation Kit (TBD, Tianjing, China, catalog # LZS11131) according to the manufacturer’s instructions. Briefly, fresh blood from healthy donors was overlaid onto the neutrophil isolation reagent. After centrifugation for 25 min at 750 × g, the neutrophil-enriched supernatant was collected. Two rounds of residual erythrocyte lysis were followed by centrifugation for 10 min at 450 × g. Neutrophils were washed twice with phosphate-buffered saline (PBS) (Bioind, Israel, catalog # 02-024-1ACS) and resuspended in Roswell Park Memorial Institute (RPMI) 1640 medium (Bioind, Israel, catalog # 01-100-1ACS) supplemented with 3% fetal bovine serum (FBS) (Bioind, Israel, catalog # 04-001-1ACS) (1 × 106 cells/ml).
Neutrophil stimulation and NET isolation
A volume corresponding to 5 × 106 neutrophils was inoculated into 25 cm2 cell culture flasks. After incubation with 500 nM PMA for 4 h in a 37°C incubator with 5% CO2, neutrophils were subjected to NET isolation protocols A-D. Each protocol was repeated at least in sextuplicate.
Quantification of NETs
A PicoGreen dsDNA kit (Thermo Fisher Scientific,Waltham, USA, catalog # P7589) was used for NET quantitation according to the manufacturer’s instructions. Briefly, a total of 100 µl of isolated NET was transferred to 96-well plates, followed by 100 µl of 1X Quant-iT Pico-Green reagent. Plates were incubated for 5 min at room temperature prior to fluorescence detection (excitation/emission wavelengths: 502/523 nm) using a SpectraMax M2 spectrofluorometer (Molecular Devices, Biberach an der Riß, Germany).
Statistical analysis
All statistical analyses were performed using SPSS Statistics 23.0 for Windows 10 (IBM, New York, U.S.). A one-way analysis of variance (ANOVA) was used to analyze the differences between groups. For multiple comparisons, Dunnett’s test was selected when one column represented control data, and the Tukey test was selected when comparing all pairs of columns. All values representing normally distributed data are expressed as the mean ± standard deviation. A p-value < 0.05 was considered statistically significant.
Results
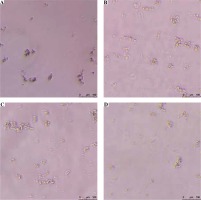
Although protocol C (Addition of the restriction enzyme AluI prior to centrifugation) is most effective, yielding a NET concentration of 1077.22 ±229.04 ng/ml, protocol B (Immediate centrifugation to pellet neutrophils) is the most efficient, with implementation times of only 15 min each (Table 2). All protocols yielded highly pure NET fractions free of neutrophil contamination (Fig. 1).
Discussion and conclusions
To the best of our knowledge, this is the first study comparing the efficacy and efficiency of different human NET isolation and handling protocols. Protocol C was found to be most effective, generating the highest NET yield. This may be attributable to the inclusion of the restriction enzyme AluI, which cleaves DNA at 5'-AG/CT-3' recognition sites to generate small NET fragments [14], thereby minimizing loss during centrifugation. In addition, because DNA fragmentation will significantly affect DNA concentration as estimated using fluorimetry, this method may significantly underestimate yields resulting from protocol C [15]. Although protocols A and D both generated highly pure NET fractions, their yields were relatively low, likely due to inclusion in these protocols of double the number of centrifugation steps relative to other protocols. In addition, both protocols A and D require access to an ultracentrifuge. Finally, it is worth noting that if pathogens replace PMA as NETosis triggers, they may prove difficult to remove via subsequent centrifugation during protocol D.
In conclusion, although protocol C produced the highest NET yield, balancing protocol efficacy and efficiency suggests that protocol B (incorporating centrifugation for 5 min at 450 × γ to pellet neutrophils) is more than adequate.