Introduction
Ankylosing spondylitis (AS) is a common inflammatory arthritis characterised by acute and chronic inflammation of the axial skeleton, causing pain, stiffness, and progressive joint ankylosis. AS has been strongly associated with human leukocyte antigen-B27 (HLA-B27), and its contribution to the genetics of AS is best characterised at present in various populations [1-4]. For instance, HLA-B27 is found in more than 80% of Chinese patients with AS [5], while only 1-5% of HLA-B27 carriers develop AS in the Chinese population [6]. However, the reported prevalence of AS ranges between 2% and 5% among major Arab populations [7]. Moreover, association studies on AS in the Maghreb population have mainly focused on the implication of the HLA-B27 gene in disease susceptibility [8-10], thus showing a weaker association between AS and HLA-B27 for Algerian patients as compared to European patients (63% vs. 80-90%, respectively) [8].
Although the contribution of HLA-B27 to the genetics of AS is best characterised at present, the pathogenesis of AS remains poorly understood, and other genetic factors encoded by non-major-histocompatibility-complex (non-MHC) genes may contribute to this disease [11, 12]. Accordingly, many previous association studies tagging a number of non-MHC candidate genes have identified significant associations with AS in the Chinese population [13-17]. Recently, a genome-wide association study in Caucasian European populations from Australia, North America, and the United Kingdom provided the first evidence that interleukin-23 (IL-23) is involved in the pathogenesis of ankylosing spondylitis, and variants in several genes involved in the IL-23 pro-inflammatory cytokine pathway have been shown to be associated with the disease [18].
Interleukin-23 and interleukin-12 are critical cytokines that bridge innate and adaptive immunities, although they have different activities. IL-23 promotes a T-cell population characterised by the production of IL-17, IL-17F, TNF, and IL-6 [19]. IL-12β is on chromosome 5q31 and encodes the IL-12 p40 subunit that heterodimerises with the IL-12 p35 subunit (IL-12A gene) to form IL-12 cytokine, or with the IL-23 p19 subunit (IL-23A gene) to form IL-23 cytokine. IL-12β is important for the differentiation of naive T cells into IFN-γ-producing T-helper 1 cells (Th1), which is essential for the antimicrobial response [20], and this could explain its role in AS and other inflammatory-mediated processes [21]. The functional roles of IL-12β polymorphisms are still unclear, but most of the case-control studies on inflammatory diseases, including ankylosing spondylitis, have analysed one important variant in the IL-12β gene: the 3’ untranslated region (3’UTR) variant rs3212227 (or A1188C) [22-24].
The aim of our work was to confirm whether there is any association between ankylosing spondylitis and four SNPs (single-nucleotide polymorphisms) within three genes (IL-12β, JAK2, and STAT3), all involved in the IL-23 signalling pathway, for two ethnically different populations: Han Chinese and Algerian. In addition, we performed for each population a gene-gene interaction analysis to determine whether these four sequence variants could have any substantial effect increasing the incidence of AS.
Material and methods
Subjects
Two different populations were included in this case-control study, Han Chinese and Algerian. Experiments were carried out separately for the two cohorts in the Institute of Medical Genetics of Shandong University. We recruited 1010 unrelated Chinese individuals (430 patients with ankylosing spondylitis and 580 healthy controls) living in the same urban area and collected from the Department of Rheumatology and Immunology of Qilu Hospital in Shandong, an eastern coastal province in the north of China. We also recruited 250 Algerian individuals (130 unrelated AS patients and 120 healthy controls) from EHS Ben Aknoun, a central hospital in Algiers, Algeria.
Ankylosing spondylitis was diagnosed according to the modified 1984 New York criteria [25]. Magnetic resonance imaging (MRI) and computerised tomography (CT) were used as imaging modalities for sacroiliitis. Diagnosis of AS was confirmed by a qualified rheumatologist. All patients were only affected by ankylosing spondylitis, while those displaying other diseases such as inflammatory bowel disease (IBD) or psoriasis were excluded.
All healthy, unrelated controls recruited for this study were matched for age. Detailed information, clinical characteristics, and original medical institution were obtained for all participants, who gave written informed consent. The study was approved by the relevant Ethics Committee.
SNP selection and genotyping
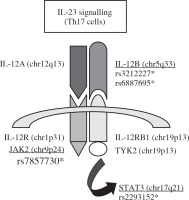
Four single-nucleotide polymorphisms (SNPs) within three genes were selected for the Han Chinese cohort from the international HapMap project database (http://www.hapmap.org/) with the Chinese Beijing Han population (CHB), namely rs3212227 (A1188C) and rs6887695 for the same gene IL-12β, rs7857730 for JAK2, and rs2293152 for STAT3. All these SNPs are involved in the IL-23 signalling pathway (Fig. 1). These four SNPs were also investigated for the first time in an Algerian cohort, since no previous association studies between AS and the three studied genes were available for the Algerian population.
Fig. 1
Schematic representation of the three candidate genes and the four tag SNPs selected for this study in the IL-23 signalling pathway (SNPs rs3212227 and rs6887695 in IL-12β, SNP rs7857730 in JAK2, and SNP rs2293152 in STAT3)
Genomic DNA from all Chinese subjects was extracted from peripheral blood mononuclear cells (PBMCs; leukocytes) by a standard phenol-chloroform method and was diluted to 20 mg/ml. QIAamp DNA Mini Kit® was used to extract genomic DNA from peripheral blood cells for all Algerian individuals [26].
Genotyping of the two cohorts involved allelic discrimination by TaqMan® SNP Genotyping Assays with an ABI prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA), using four pre-made, double-dye probes (C_2084293_10 for rs3212227), (C_1994992_10 for rs6887695), (C_3140302_10 for rs2293152), and (C_29340600_20 for rs7857730). The endpoint fluorescence was read using the Roche 480 Real-Time PCR System (Hoffmann-La Roche Ltd, Basel City, Switzerland) with 96-well plates. The PCR reaction was run in a total solution volume of 5 µl containing 2.5 µl of 2 × c Premix EX Taq (Takara, Otsu, Shiga, Japan), at least 10 ng of genomic DNA, 2.5 µM of each primer, and 0.05 µl of probe. The PCR amplification conditions were 95°C for 30 sec, 45 cycles of 95°C for 5 sec, 60°C for 20 sec, and 40°C for 30 sec for cooling. Genotyping accuracy in the samples was confirmed by direct sequencing of PCR products for 5% of randomly chosen samples. Only those individuals with 100% genotype success for all markers were included for final analysis.
Statistical analysis
Single nucleotide polymorphisms were assessed for genotypic and allelic association analysis among cases and controls, separately for the Chinese and the Algerian cohorts. The genotypic data of the four SNPs were tested for Hardy-Weinberg equilibrium by comparing genotype frequencies within the two groups (AS patients and controls) by Chi-squared test. The odds ratios (ORs) with 95% confidence intervals (95% CIs) were also calculated for single site analysis to estimate the risk associated with ankylosing spondylitis. Statistical analyses were carried out to calculate p-values using SPSS statistical software (SPSS Statistics 16.0; 2008 SPSS Inc., Chicago, IL).
In order to analyse gene-gene interactions, multiplicative interactions were estimated separately for the two cohorts using SPSS 16.0 software for logistic regression analyses. The multiplicative variable model was defined as disease status (1 is unaffected, 2 is affected) and SNP genotypic values ranging from 0 to 2 indicating the number of risk alleles in an individual subject (0 is zero risk allele, 1 is one risk allele, 2 is two risk alleles). For each SNP, random combinations were constructed to estimate a binary logistic regression model based on the case-control status (dependent variable) and the other SNP variables studied (independent variables or covariates).
Because the effect of any single genetic variation was likely to be dependent on other genetic variations, multifactor dimensionality reduction (MDR) software analysis (http://sourceforge.net/projects/mdr/) was performed to test various interaction models for one-variable, two-variable, three-variable, and four-variable gene-gene epistasis on ankylosing spondylitis. We analysed all data using 10-fold cross-validation.
Results
Association results for the Han Chinese cohort
Genotypic and allelic association analysis of Chinese cases and controls is shown in Table 1. The frequency distributions of the four SNPs within patients and controls were in Hardy-Weinberg equilibrium (p > 0.05). For the Chinese Han cohort, the SNP IL-12β rs3212227 (or A1188C) showed the strongest association with AS (p genotype = 0.007, p allele = 0.005 [OR 0.77, 95% CI: 0.65-0.93]). Another association with AS was also observed for SNP rs7857730 of the JAK2 gene (genotypic p = 0.029 and allelic p = 0.014). The minor allele frequency results revealed that C and T might be protective alleles for rs3212227 and rs7857730, respectively. Thus, subjects carrying A and G alleles showed increased risk of AS for the same variants. However, no significant differences were observed between AS patients and controls for the two other variants (IL-12β rs6887695 and STAT3 rs2293152) within the Han Chinese cohort.
Table 1
Genotypic and allelic association analysis of the four single-nucleotide polymorphisms in the Han Chinese and Algerian ankylosing spondylitis (AS) cohorts
Association results for the Algerian cohort
Association analysis results of Algerian patients and controls are represented in Table 1. All calculated frequencies were in Hardy-Weinberg equilibrium. Two variants showed associations with AS: rs3212227 of IL-12β (allelic p = 0.031) and rs2293152 of STAT3 (allelic p = 0.006). These results might reveal that A and C represent risk alleles for Algerian AS patients within the IL-12β and STAT3 genes, respectively. For variants IL-12β rs6887695 and JAK2 rs7857730, no significant association was found between AS and Algerian individuals.
Multiplicative interaction analysis results for the two cohorts
The analysis of random combinations of two, three, or four SNPs by binary logistic regression model revealed 11 interactions for each cohort, as summarised in Table 2. Significantly different results between Chinese AS patients and controls were shown by the combinations rs3212227 * rs6887695 (p = 0.008), rs6887695 * rs7857730 * rs2293152 (p = 0.006), and rs3212227 * rs6887695 * rs7857730 * rs2293152 (p = 0.045). For Algerian AS patients and controls, only rs3212227 * rs7857730 * rs2293152 was found as a significant combination (p = 0.035).
Table 2
Multiplicative interaction analysis reported by logistic regression for the two populations investigated
MDR analysis results and gene-gene epistasis for the two cohorts
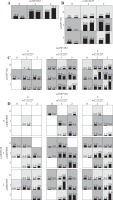
Various interaction models were tested by MDR analysis in order to detect any potential interaction effect of gene-gene epistasis represented in Table 3 for the two cohorts. All data were based on the highest testing accuracy and cross-validation consistency. For the Han Chinese cohort, the one-variable model SNP IL-12β rs3212227 showed the highest testing accuracy among the four SNPs. The two-variable gene-gene interaction model (IL-12β rs3212227, JAK2 rs7857730) and the three-variable gene-gene interaction model (IL-12β rs3212227, JAK2 rs7857730, and STAT3 rs2293152) showed the highest testing accuracy as compared to the other two-locus and three-locus interaction models (p = 0.0001 and p < 0.0001; respectively). MDR also showed a high testing accuracy for the four-variable gene-gene interaction model (p < 0.0001) for the Chinese cohort (Fig. 2).
Table 3
Gene-gene interaction model results of MDR for each population
Fig. 2
Graphic models determined by multifactor dimensionality reduction (MDR), representing the following different interactions in the Chinese cohort: A) IL-12β rs3212227, B) IL-12β rs3212227 and JAK2 rs7857730, C) IL-12β rs3212227, JAK2 rs7857730, and STAT3 rs2293152, D) IL-12β rs3212227, IL-12β rs6887695, JAK2 rs7857730, and STAT3 rs2293152. SNP genotypic values range from 0 to 2 (0 = zero risk allele, 1 = one risk allele, 2 = two risk alleles). Each cell shows counts of “Class 1 = unaffected” on the left and “Class 2 = affected” on the right, and shades on cells represent the risk degree of the disease (dark cell = high risk, light cell = low risk)

For the Algerian cohort, the one-locus model SNP STAT3 rs2293152 represented the model with highest testing accuracy among the four SNPs. The two-locus (IL-12β rs3212227 and JAK2 rs7857730), the three-locus (IL-12β rs3212227, JAK2 rs7857730, and STAT3 rs2293152), and the four-locus interaction models all showed the highest testing accuracy (p < 0.0001) within the other interaction models (Fig. 3).
Fig. 3
Graphic models determined by multifactor dimensionality reduction (MDR), representing the following different interactions in the Algerian cohort: A) STAT3 rs2293152, B) IL-12β rs3212227 and JAK2 rs7857730, C) IL-12β rs3212227, JAK2 rs7857730, and STAT3 rs2293152, D) IL-12β rs3212227, IL-12β rs6w887695, JAK2 rs7857730, and STAT3 rs2293152. SNP genotypic values range from 0 to 2 (0 = zero risk allele, 1 = one risk allele, 2 = two risk alleles). Each cell shows counts of “Class 1 = unaffected” on the left and “Class 2 = affected” on the right, and shades on cells represent the risk degree of the disease (dark cell = high risk, light cell = low risk)
Discussion
While the precise molecular aetiology of ankylosing spondylitis remains unclear, a variety of genetic factors have been involved in the development and incidence of the disease. Thus, many studies have reported a number of new non-MHC genes to be involved in the pathogenesis of AS, in particular those involved in the IL-23 pathway [27-29]. In the present study, our data indicated that SNP IL-12β rs3212227 (+1188A/C) was significantly associated with AS in the Han Chinese cohort. This polymorphism was also associated with AS in the Algerian cohort, and it is worth noting that the AA genotype may increase the risk of AS in both Chinese and Algerian subjects. Previously, linkage analysis studies in the Chinese population have considered IL-12B as a candidate gene for AS, and several polymorphisms were tested within this gene, but their results were contentious regarding whether some of these polymorphisms are associated with AS susceptibility and others are associated with disease severity [30, 31]. Nevertheless, in a recent association study of IL-12B polymorphism susceptibility with ankylosing spondylitis in the Mainland Han population, no significant difference was found for IL-12β rs3212227 variant, while only the SNP rs6871626 from the eight polymorphisms studied showed a significant difference in genotype distribution between AS and healthy controls [32]. Another meta-analysis studying the relationship between rs3212227 polymorphism of the IL-12B gene and susceptibility to multiple autoimmune diseases in East Asian populations failed to find any association with Graves’ disease and ankylosing spondylitis [23]. Additionally, the International Genetics of Ankylosing Spondylitis Consortium (IGAS) reported other SNPs within IL-12β that conferred risk of ankylosing spondylitis in Europeans, but the two SNPs analysed in our study were not genotyped [28].
In fact, IL-12β may play a key role in the pathogenesis of several inflammatory diseases because it encodes the p40 subunit of IL-23 and IL-12 cytokines and is thus involved in both the IL-12/Th1 and IL-23/Th17 pathways. Many studies were recently conducted to confirm the susceptibility of polymorphisms in IL-12β, in particular rs3212227 (+1188A/C) polymorphism, to cause ankylosing spondylitis (AS) and/or other autoimmune diseases such as rheumatoid arthritis (RA), type 1 diabetes (T1D), Behcet’s disease (BD), Graves’ disease (GD), and multiple sclerosis (MS) [23, 33]. Hence, taken together, our results may accentuate the importance of IL-12β rs3212227 (+1188A/C) as a novel risk polymorphism in the development of ankylosing spondylitis in both Chinese and Algerian subjects. Unfortunately, the second polymorphism in IL-12β rs6887695 was not associated with susceptibility to AS, neither in the Chinese nor in the Algerian individuals. The two polymorphisms rs3212227 and rs6887695 in IL-12β have been previously found to be associated with risk of psoriasis and psoriatic arthritis in the Chinese population [24, 34].
We further analysed the two other polymorphisms rs2293152 in STAT3 and rs7857730 in JAK2, but our results were only partially in agreement with the findings of Chen et al. [14], who reported no difference in the distribution of alleles and genotypes between AS groups and controls in their Chinese cohort in both JAK2 and STAT3 genes. In fact, our findings only showed a significant association between Chinese AS patients and controls for rs7857730 in the JAK2 gene. The difference in these findings is probably due to the difference in the recruitment of Chinese subjects for the two studies. Chen et al. also reported that haplotype analysis revealed an association of haplotype rs1536798/rs10119004/rs7857730-CGT in the JAK2 locus with AS, which is consistent with our results. Therefore, our findings may confirm the association of rs7857730 polymorphism in JAK2 with the susceptibility to AS and may emphasise again the importance of the IL-23/IL-17 axis as a causal pathway of AS in the Chinese population, for which earlier studies have also investigated its role in AS [27] and related diseases such as IBD [35] and psoriasis [36].
Another case-control study, carried out by Davidson et al. [15], in which a number of genes implicated in the pathogenic mechanisms of AS were tagged in the Han Chinese population, showed that SNP rs2293152 in STAT3 had a significant association with AS in their Chinese cohort, a result that corroborates the one previously found in STAT3 rs2293152 variant by the Australo-Anglo-American Spondyloarthritis Consortium (TASC) in Caucasian European populations [18]; however, this was in contrast to our results.
Unlike the Chinese cohort, we reported no significant difference between Algerian AS patients and controls for the variant JAK2 rs7857730, but a significant association for the variant STAT3 rs2293152 was found in this group. This discrepancy in our findings could be explained by differences in the genetic pools between the two populations investigated.
Our analysis also considered different interactions among one-variable, two-variable, three-variable, and four-variable gene-gene epistasis with ankylosing spondylitis, which were analysed to address concerns about DNA sequence variations that could have an interaction effect but not a statistically significant independent main effect on AS. Remarkably, logistic regression analysis showed significant interactions between IL-12β rs6887695 and the three other variables in the Han Chinese cohort, although this SNP itself showed no significant association with AS. MDR analysis also revealed a gene-gene interaction model for the Han Chinese cohort, in which IL-12β rs6887695 interacts with the other variables. This difference for IL-12β rs6887695 between individual association results and multiple interaction findings could be explained by the relatively small sample size. In addition, IL-12β rs3212227 showed the strongest association with AS in the Chinese population, so the significant interaction observed between the two variables of the IL-12β gene (rs3212227 and rs6887695) could confirm once again the important role played by this gene in the IL-23 signalling pathway and the pathogenesis of AS.
Analysis of interaction by logistic regression in the Algerian cohort revealed a significant three-variable interaction between IL-12β (rs3212227), JAK2 (rs7857730), and STAT3 (rs2293152) SNPs for AS susceptibility (p = 0.035). Because both IL-12β and STAT3 polymorphisms individually analysed showed a significant association with AS, this interaction suggests that IL-12β and STAT3 may be involved in the risk of developing AS in Algerian patients. In fact, STAT3 is predominantly activated by IL-23, which signals through IL-12Rβ1 and IL-23R to activate JAK and STAT signalling molecules, and induces IL-17A, IL-17F, and/or IL-22 and stabilises TH17 cells [19]. Thus, the additive interaction effect of the two SNPs in STAT3 and IL-12β may increase the risk of AS for Algerian patients.
In conclusion, we confirmed in our study a significant association between ankylosing spondylitis and the IL-12β gene for the rs3212227 variant in both the Han Chinese and Algerian populations. Also, JAK2 rs7857730 polymorphism showed an association in the Han Chinese population, while rs2293152 polymorphism in the STAT3 gene was associated with AS in the Algerian population. Despite the relatively small sample sizes and the number of SNPs selected for each gene, our findings remain novel. Additionally, the gene-gene interaction analysis through the effect of different SNPs was combined and may therefore provide a better understanding of AS and its pathogenic mechanisms.


