Introduction
Hand, foot and mouth disease (HFMD) is a major threat to the health of infants [1-5]. In 2008, HFMD was listed as a class C infectious disease (which is mandatory to be reported) in the “People’s Republic of China Disease Prevention Act”. The pathogens that cause HFMD are mainly enterovirus 71 (EV71) and Coxsackie virus type A16 (CA16) [6-8]. CA16 also causes myocarditis, cardiomyopathy, pericarditis and other serious diseases, which accounts for more than 31.9% of the total number of infections [9]. CA16 was first isolated in Canada in 1958. CA16 was classified as a small RNA virus family (Picornaradae) and enterovirus genus (Enterovirus). The structural proteins of CA16 include VP1, VP2, VP3, and VP4, of which VP1, VP2, and VP3 were present on the viral surface, while VP4 is located near RNA. Although extensive studies on CA16 have been reported, effective therapeutic drugs and vaccines are still not available. Thus, it is difficult to prevent and treat CA16 infections by vaccines and drugs that are developed by conventional methods. In this study, we fused VP1 gene with CotB and introduced this recombinant gene into the genome of Bacillus subtilis. VP1 was displayed on the surface of Bacillus spores. This Bacillus-based mucosal vaccine could induce strong neutralizing antibody response in mice and thus could potentially be used to protect from CA16 infection.
Material and methods
This study was approved by the institutional review board of Liaocheng People’s Hospital.
Plasmid pDG1662 and Bacillus subtilis 1A771 were purchased from Bacillus Genetic Stock Center (Columbus, USA). CA16 was isolated from the stool specimens of HFMD patients admitted to Liaocheng People’s Hospital after obtaining written consent. BALB/c mice were purchased from the Experimental Animal Center of Shandong University (Jinan, China). Cloning vector pMD18-T and pfu DNA polymerase were purchased from TaKaRa Biotechnology (Dalian, China). The RNA extraction kit, DNA gel extraction kit and plasmid extraction kit were purchased from Tiangen Technology (Beijing, China). The restriction enzyme and reverse transcriptase kit were purchased from Thermo (Waltham, MA USA). HRP conjugated goat anti-mouse IgG was purchased from Beijing Biosynthesis Biotechnology (Beijing, China). HRP conjugated goat anti-mouse IgA was purchased from Abcam (Cambridge, UK). FITC-conjugated goat anti-mouse IgG was purchased from Wuhan Boster Biological Engineering (Wuhan, China). Complete Freund’s adjuvant and incomplete Freund’s adjuvant were purchased from Sigma (Shanghai, China). Chemical reagents were purchased from Sinopharm Chemical Reagent (Shanghai, China).
PCR primers
CotB and VP1 genes were amplified by the following primers:
CotB1: 5’-GAATTCGAATCCGAGTTTCGCAAGTCCT-3’;
CotB2: 5’-AAGCTTGATGATTGATCATCTGAAGAT-3’ (expected size: 1305 bp);
VP1.1: 5’-AAGCTTGGGGACCCCATTGCAGATATGAT-3’;
VP1.2: 5’-GGATCCCAGCGTTGTTATCTTGTCTCTACT-3’ (expected size: 903 bp).
Primers were synthesized by Shanghai Boya Bio Co (China). RNA Extraction Kit was used to isolate CA16 genome and Oligo (dT) was used as the primer to obtain cDNA by the reverse transcription. cDNA and Bacillus 1A771 DNA were used as templates to amplify the VP1 and CotB genes, respectively. The amplified polymerase chain reaction (PCR) products were ligated into pMD18-T vector (resulting in the plasmids pMD18-VP1 and pMD18-CotB, respectively) and confirmed by DNA sequencing.
Construction of recombinant plasmids
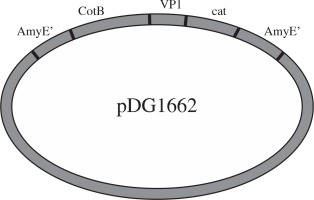
Plasmids pMD18-VP1 and pMD18-CotB were double digested by restriction enzyme pair of EcoRI/HindIII and HindIII/BamHI, respectively. The digested fragments were ligated into pDG1662 that was digested with the EcoRI and BamHI (Fig. 1). The resulting plasmids containing VP1 and CotB fusion gene were verified by PCR using CotB1-CotB2, VP1.1-VP1.2 and CotB1-VP1.2 primers and restriction digestion using EcoRI/HindIII, HindIII/BamHI and EcoR/BamHI.
Transformation and screening
A loop of 1A771 spores were inoculated into 5 ml medium and cultured overnight. The overnight culture was inoculated into 80 ml growth medium and incubated for 3 hours with shaking. The culture was incubated on ice for 10 min and centrifuged (4oC, 5000 × g) to collect the bacterial pellet. Competent cells were prepared by washing the Bacillus 3 times using cold electroporation buffer. Plasmid pDG1662-CotB-VP1 was linearized by restriction enzyme NdeI and incubated with the competent cells for 1.5 min. Following electroporation (1800 V), 1 ml recovering medium was added to the cuvette and cultured for 3 hours. The culture was spread on LB plate with chloramphenicol. Colonies that are resistant to chloramphenicol, but sensitive to erythromycin, were the correct recombinant strain.
Verification of the recombinant strain by PCR
Colonies that were resistant to chloramphenicol, but sensitive to erythromycin, were cultured in 10 ml LB containing chloramphenicol overnight. Genomic DNA was extracted by phenol chloroform. VP1 primers were used to verify if the recombinant strains were correctly constructed.
Verification of the recombinant strain by Western blot
Chloramphenicol resistant strain was cultured in LB at 37oC with shaking for 24 hours followed by static standing at room temperature for 48 hours to generate sufficient spores. Cells were collected by centrifugation (10 000 × g) and treated with lysozyme (20 mmol/l Tris-HCl buffer solution, pH 8.0, lysozyme 1 g/l) at 37oC for 1 hour. Spores were collected by centrifugation. Shelling buffer (10 ml) was used to resuspended spores. Spores were incubated at 70oC with 250 rpm shaking for 1 hour. Following centrifugation at 14 000 × g, the supernatant contains capsid protein. Wild-type strain was prepared similarly. Capsid protein and 2 × sample buffer was mixed and boiled for 3 min. After SDS-PAGE and transferring to nitrocellulose membrane, western blot was performed using polyclonal anti-CA16 as the primary antibody and HRP conjugated goat anti-mouse IgG as the secondary antibody.
Characterization of the recombination strain by fluorescent microscopy
Spores were washed with phosphate-buffered saline (PBS) for 3 times and incubated with CA16 polyclonal antibody overnight. Following 3 times of washing with PBST, the spores were resuspended and incubated with FITC-conjugated goat anti-mice IgG for 1 hour at 37oC with slight shaking. Subsequently, the spores were washed and placed on the glass slide for fluorescent microscopic analysis.
Animal immunization
A total of 26 BALB/c mice were randomly divided into experimental and control groups (n = 10 per group) and the remaining 6 mice were used as a negative group. The experimental, control and negative groups were inoculated with the recombinant Bacillus, VP1 protein and saline solution, respectively, via intranasal immunization twice a week for a total of 6 weeks. At week 7, blood sample was collected from ophthalmic artery plexus and the serum was stored at –80oC for later use.
Measurement of antibody titer by ELISA
The 96-well plate was coated with inactivated CA16 virus and blocked by 5% non-fat milk. The serum was 2-fold diluted with PBS and added into the each well (100 ml/well). After incubation at 37oC for 1 hour, HRP-conjugated goat anti-mouse immunoglobulin (Ig) G and IgA was added. Following additional 1 hour’s incubation at 37oC, TMB substrate was added. A450 was measured after the reaction was terminated by sulfuric acid (2M). SPSS13.0 was used to analyze the antibody titer in the serum of mice.
Neutralization test
Neutralization test was performed as described previously [10-12]. Briefly, mouse serum was diluted at 1 : 10 and inactivated at 56°C for 30 minutes. Inactivated serum was serially diluted at 1 : 10. Serum at each dilution was added with an equal volume of virus (400 TCID50) and incubated for 1 hour at 37oC. Each dilution was then inoculated onto 96-well plate (50 ml/well). Each well was also added with 50 ml (1 × 104) RD cells. Following culture at 37oC and 5% CO2 condition, pathological alterations of the cells were determined. The virus control, and normal serum control and normal cell control were included. The highest dilution without pathological alteration was considered as the neutralizing antibody titers. SPSS13.0 was used for data analysis.
Results
Amplification of the VP1 and CotB genes
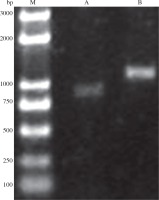
PCR results showed that CotB and VP1 genes were successfully amplified with an expected size of 1305 and 904 bp, respectively (Fig. 2). Sequence analysis showed that CotB and VP1 genes were inserted into pMD18-T vector, resulting in the plasmids pMD18-VP1 and pMD18-CotB, respectively.
Construction of the recombinant plasmid
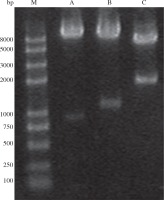
Plasmids pMD18-VP1 and pMD18-CotB were double digested with the restriction enzyme pair of EcoR/HindIII and HindIII/BamHI, respectively. The fragments (CotB and VP1) excised from the two plasmids were inserted into pDG1662, resulting in the recombinant plasmid pDG1662-CotB-VP1. Restriction digestion of pDG1662-CotB-VP1 with EcoR/HindIII, HindIII/BamHI and EcoR/BamHI resulted in the expected bands of 1305bp, 904bp and 2209bp, respectively (Fig. 3), which indicated that CotB and VP1 fusion was constructed into pDG1662 plasmid.
Characterization of the recombinant strain by PCR
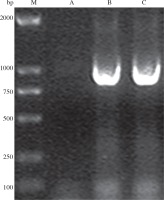
PCR using genomic DNA extracted from the recombinant strain as template and VP1.1-VP1.2 as primers resulted in an expected size of 904 bp band, while the control did not have positive band (Fig. 4), which indicated that VP1 was successfully inserted into the genome of Bacillus subtilis.
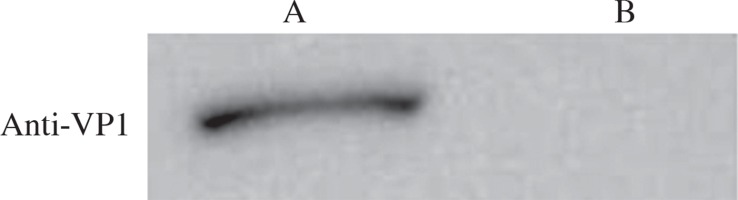
Characterization of the recombinant strain by Western blot
Proteins were extracted from wild-type and recombinant Bacillus subtilis spores and resolved by SDS-PAGE. Western blot was performed using VP1 polyclonal antibody as the primary antibody and HRP-conjugated goat anti-mouse IgG as the secondary antibody. The results showed a positive band in the recombinant strain, but not in the wild-type strain (Fig. 5), which indicated that VP1 was expressed in the spores of the recombinant strain.
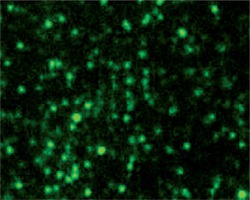
Characterization of the recombinant strain by immunofluorescence microscopy
The spores of the recombinant and wild-type strains were incubated with VP1 polyclonal antibody and FITC-conjugated goat anti-mouse IgG. Fluorescence microscopy showed that the recombinant strain exhibited green fluorescence (Fig. 6), while the wild-type strain did not exhibit green fluorescence (Fig. 6). These results indicated that VP1 was displayed on the surface of spores and had strong immunogenicity.
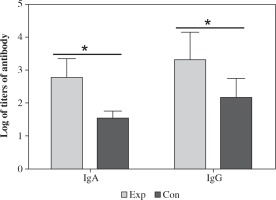
Antibody titers measured by ELISA
Mice were immunized with the recombinant strain and the antibody including IgG and IgA in the serum was measured by ELISA using CA16 as antigen. The results showed that CA16 specific antibody was induced in mice. The highest and lowest titer of IgA was 1 : 1280 and 1 : 320 (geometric mean titer was 1 : 597), respectively.The highest and lowest titer of IgG was 1 : 5120 and 1 : 640 (geometric mean titer was 1 : 2087), respectively (Fig. 7). The antibody titers in the mice immunized with the recombinant strain was significantly higher than those immunized with VP1, which suggested that intranasal immunization induced specific humoral immune response. The spores can significantly increase the antibody level and can function as an excellent mucosal adjuvant.
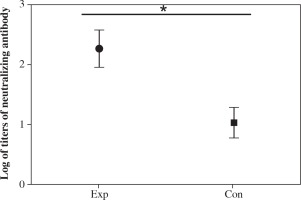
Cell neutralizing assay
Serially diluted serum from each group of mice was used for neutralizing assay. The results showed that the highest and lowest titer of the antibody from the recombinant strain group was 1 : 512 and 1 : 64 (geometric mean titer was 1 : 186), respectively (Fig. 8). The titer in the recombinant strain group was significantly higher than that in the VP1 control group (Fig. 8), which indicated that immunization with spore not only increases the ELISA titer, but also enhances the neutralizing antibody titers.
Discussion
CA16 is one of the main pathogens causing HFMD in mainland China [13-15]. Extensive studies on CA16 vaccine have been reported. Virus-like particle and inactivated virus can protect suckling mice from lethal doses of virus attacks [16, 17]. The subunit vaccines study mainly focused on VP1 capsid protein. It is well known that CA16 capsid protein is composed of VP1-4, of which protein VP1 is the major capsid protein and has a specific neutralizing epitope. Immunization of animals with VP1 can induce strong humoral and cellular immune response [18]. Thus, VP1 is the best choice for preparing CA16 subunit vaccine. However, whole virus or subunit vaccine researches are still in the experimental stage and alternative approaches are needed to develop an ideal CA16 vaccine. IgA is one of the important immunoglobulins. IgA is particularly important in resistance to pathogenic microorganisms, especially those enteric viruses that cause infections via the mucosal route. IgA is thought to protect the host against pathogens that infect mucosal surfaces and those that cause systemic disease after entry via a mucosal surface [19]. Therefore, induction of high levels of IgA and IgG antibodies through the mucosal immunization route may provide new strategies to prevent and control CA16 infection. It has been reported that EV71 vaccine prepared by displaying VP1 protein on baculovirus can effectively induce immune response in mice [20]. Bacterial surface display technology, which is similar to the baculovirus display, also received widespread attention in the field of vaccine. Extensive studies using spore as the vaccine vector have been reported [21-23]. The spore vector is safe, stable, easy to prepare, and becomes a potentially effective tool for vector vaccine. Many studies have shown that mucosal subunit vaccine VA16 VP1 using spore as a vector is feasible.
In this study, we amplified VP1 gene from CA16 and inserted VP1 in fusion with CotB gene from Bacillus subtilis into pDG1662 plasmid. VP1 was inserted into the genome of Bacillus via recombination. Erythromycin sensitive and chloramphenicol resistant clones were selected. During the process of screening, we found that not all chloramphenicol resistant clones were sensitive to erythromycin, which suggested that erythromycin resistant gene was not excised during the recombination. It also indicated that there were different ways of integration of pDG1662 into the genome. PCR, Western blot and immunofluorescence microscopy analysis demonstrated that VP1 gene was successfully integrated into the genome of Bacillus subtilis and was expressed on the surface of Bacillus spores. However, it is worth noting that the band in Western blot was weak, which indicated that VP1 expression was low, which is consistent with previous reports [24].
Intranasal immunization of mice with this vaccine induces a significantly higher level of VP1 specific IgA and IgG compared the non-immunized group, which suggested that mucosal immunization of VP1 using spore as adjuvant can enhance immunization efficacy and humoral immune response. More importantly, the neutralizing antibody titer in the spore immunization group was 1 : 169, which was significantly higher than that in the control group. These results indicated that mucosal vaccine using spore as a vector can increase not only the titer of the specific antibody, but also the titer of the neutralizing antibody.
In summary, this study reported, for the first time, the CA16 VP1 subunit vaccine using spore as the expression system and adjuvant. This vaccine can effectively stimulate a specific neutralizing antibody via intranasal immunization and exhibit virus inhibitory activities. Our study provides new strategies to develop CA16 vaccine and sets the foundation for CA16 mucosal vaccine. This work represents an advance in biomedical science because it is introducing a new vaccine for the prevention of CA16 infection for the first time.